Volume 19, Issue 4 (Jul-Aug 2025)
mljgoums 2025, 19(4): 1-4 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Hasan S A, Raoof W M, Ahmed K K. Prevalence and antibiotic resistance patterns of carbapenemase-producing organisms in burn and wound infections. mljgoums 2025; 19 (4) :1-4
URL: http://mlj.goums.ac.ir/article-1-1805-en.html
URL: http://mlj.goums.ac.ir/article-1-1805-en.html
1- College of Medicinal and industrial plants, University of Kirkuk, Kirkuk, Iraq , sarahahmed100@uokirkuk.edu.iq
2- Biology Department, Science College, Tikrit University, Tikrit, Iraq
3- College of Medicinal and industrial plants, University of Kirkuk, Kirkuk, Iraq
2- Biology Department, Science College, Tikrit University, Tikrit, Iraq
3- College of Medicinal and industrial plants, University of Kirkuk, Kirkuk, Iraq
Keywords: Carbapenemase, Pseudomonas aeruginosa, Escherichia coli, Enterobacter cloacae, Klebsiella pneumoniae
Full-Text [PDF 462 kb]
(1102 Downloads)
| Abstract (HTML) (4435 Views)
Full-Text: (872 Views)
Introduction
Carbapenems are β-lactam antibiotics that have multiple bactericidal activities (1). These antibiotics are usually considered the last-choice drugs in treating infections with multidrug-resistant (MDR) Gram-negative bacteria. However, according to Patel and Bonomo (2011), (2) reported that carbapenemase-producing organisms (CPOs) are associated with serious healthcare-associated infections, making the mortality rate higher. Some of these bacteria contribute significantly to the prevention and treatment of infections.
The issue of carbapenem-resistant pathogens, which is a global concern these days, requires appropriate strategies at the national and international levels (3). The basic reason for carbapenem resistance is mostly achieved by the synthesis of carbapenemase enzymes, which hydrolyse carbapenem drugs (A group of β-lactam antibiotics) (4). Different genes are involved in carbapenem resistance via the production of carbapenemases. As a result, those genes that code for carbapenemases are associated with various types of mobile genetic elements (5).
Different kinds of β-lactamases hydrolysis carbapenem like metallo-β-lactamases (MBLs) which include: β-lactamase of New Delhi metal (NDM), Verona imipenemase (VIM) and Impipenemase (IMP); in addition to class A Ambler member, Klebsiella pneumoniae carbapenemase (KPC), and class D member, oxacillinase-48 (OXA-48) (6).
The transmissible carbapenemase genes in Gram negative bacteria was considered as the greatest threat to the public health across the globe, due to the fact that these pathogens limit the effective antibiotic treatments and cause high mortality rate among patients (7), these carbapenemase genes are transmitted, not only among Acinetobacter baumannii but also other nosocomial pathogens like Enterobacteriaceae family members or Pseudomonaerous aeruginosa (8).
The world faces an emergency that demands the development of new antimicrobial agents which can be applied against CPOs with MDR bacteria pattern. This study is the first in Iraq to determine the prevalence and multidrug resistance patterns of CPOs isolated from burn and wound infections.
Methods
Sample collection and culturing
A total of 250 clinical samples (140 wound swabs and 110 burn swabs) were collected aseptically from hospitalized patients in Kirkuk and Sulaimaniyah hospitals between January and July 2023. All samples were immediately transported to the microbiology laboratory under sterile conditions to prevent contamination. Upon arrival, specimens were cultured on MacConkey agar for Gram-negative selection and cetrimide agar for Pseudomonas isolation, followed by incubation at 37°C for 18-24 hours. Colony morphology, including color, shape, and texture was examined for preliminary identification.
Bacterial identification and antimicrobial susceptibility testing
Bacterial isolates were identified using the BD Phoenix™ M50 automated system. Antimicrobial susceptibility testing was performed against 18 antibiotics representing seven classes: aminoglycosides (Amikacin, gentamicin), carbapenems (Ertapenem, imipenem, meropenem), cephalosporins (Cefazolin, cefuroxime, ceftazidime, ceftriaxone, cefepime), beta-lactam combinations (Ceftolozane-tazobactam, amoxicillin-clavulanate, piperacillin-tazobactam), penicillins (Ampicillin), fluoroquinolones (Ciprofloxacin, levofloxacin), and other agents (Trimethoprim-sulfamethoxazole, tigecycline). Testing procedures followed the manufacturer's standardized protocols.
Carbapenemase detection
Carbapenemase production was detected phenotypically using the BD RAPIDEC® CARBA NP assay according to the manufacturer's instructions. This chromogenic test specifically identifies carbapenemase activity in Gram-negative bacterial isolates.
Statistical analysis
Data analysis was performed using Microsoft Excel 2016 to determine prevalence rates and resistance patterns.
Results
From the 250 clinical samples analyzed, 71 (28.4%) grew Gram-negative bacterial isolates. The distribution of pathogens was as follows: 31 isolates (43.66%) were Pseudomonas aeruginosa, 16 isolates (22.53%) were Escherichia coli, 13 isolates (18.3%) were Enterobacter cloacae, and 11 isolates (15.49%) were Klebsiella pneumoniae (Figure 1).
Carbapenems are β-lactam antibiotics that have multiple bactericidal activities (1). These antibiotics are usually considered the last-choice drugs in treating infections with multidrug-resistant (MDR) Gram-negative bacteria. However, according to Patel and Bonomo (2011), (2) reported that carbapenemase-producing organisms (CPOs) are associated with serious healthcare-associated infections, making the mortality rate higher. Some of these bacteria contribute significantly to the prevention and treatment of infections.
The issue of carbapenem-resistant pathogens, which is a global concern these days, requires appropriate strategies at the national and international levels (3). The basic reason for carbapenem resistance is mostly achieved by the synthesis of carbapenemase enzymes, which hydrolyse carbapenem drugs (A group of β-lactam antibiotics) (4). Different genes are involved in carbapenem resistance via the production of carbapenemases. As a result, those genes that code for carbapenemases are associated with various types of mobile genetic elements (5).
Different kinds of β-lactamases hydrolysis carbapenem like metallo-β-lactamases (MBLs) which include: β-lactamase of New Delhi metal (NDM), Verona imipenemase (VIM) and Impipenemase (IMP); in addition to class A Ambler member, Klebsiella pneumoniae carbapenemase (KPC), and class D member, oxacillinase-48 (OXA-48) (6).
The transmissible carbapenemase genes in Gram negative bacteria was considered as the greatest threat to the public health across the globe, due to the fact that these pathogens limit the effective antibiotic treatments and cause high mortality rate among patients (7), these carbapenemase genes are transmitted, not only among Acinetobacter baumannii but also other nosocomial pathogens like Enterobacteriaceae family members or Pseudomonaerous aeruginosa (8).
The world faces an emergency that demands the development of new antimicrobial agents which can be applied against CPOs with MDR bacteria pattern. This study is the first in Iraq to determine the prevalence and multidrug resistance patterns of CPOs isolated from burn and wound infections.
Methods
Sample collection and culturing
A total of 250 clinical samples (140 wound swabs and 110 burn swabs) were collected aseptically from hospitalized patients in Kirkuk and Sulaimaniyah hospitals between January and July 2023. All samples were immediately transported to the microbiology laboratory under sterile conditions to prevent contamination. Upon arrival, specimens were cultured on MacConkey agar for Gram-negative selection and cetrimide agar for Pseudomonas isolation, followed by incubation at 37°C for 18-24 hours. Colony morphology, including color, shape, and texture was examined for preliminary identification.
Bacterial identification and antimicrobial susceptibility testing
Bacterial isolates were identified using the BD Phoenix™ M50 automated system. Antimicrobial susceptibility testing was performed against 18 antibiotics representing seven classes: aminoglycosides (Amikacin, gentamicin), carbapenems (Ertapenem, imipenem, meropenem), cephalosporins (Cefazolin, cefuroxime, ceftazidime, ceftriaxone, cefepime), beta-lactam combinations (Ceftolozane-tazobactam, amoxicillin-clavulanate, piperacillin-tazobactam), penicillins (Ampicillin), fluoroquinolones (Ciprofloxacin, levofloxacin), and other agents (Trimethoprim-sulfamethoxazole, tigecycline). Testing procedures followed the manufacturer's standardized protocols.
Carbapenemase detection
Carbapenemase production was detected phenotypically using the BD RAPIDEC® CARBA NP assay according to the manufacturer's instructions. This chromogenic test specifically identifies carbapenemase activity in Gram-negative bacterial isolates.
Statistical analysis
Data analysis was performed using Microsoft Excel 2016 to determine prevalence rates and resistance patterns.
Results
From the 250 clinical samples analyzed, 71 (28.4%) grew Gram-negative bacterial isolates. The distribution of pathogens was as follows: 31 isolates (43.66%) were Pseudomonas aeruginosa, 16 isolates (22.53%) were Escherichia coli, 13 isolates (18.3%) were Enterobacter cloacae, and 11 isolates (15.49%) were Klebsiella pneumoniae (Figure 1).
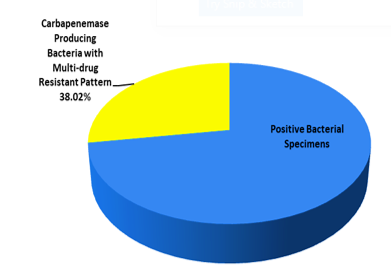 Figure 1. Prevalence of carbapenemase producing bacteria with multi-drug resistant pattern among the positive bacterial isolates |
|
Table 1. Carbapenemase-producing Gram-negative isolates and their multi-drug resistant patterns
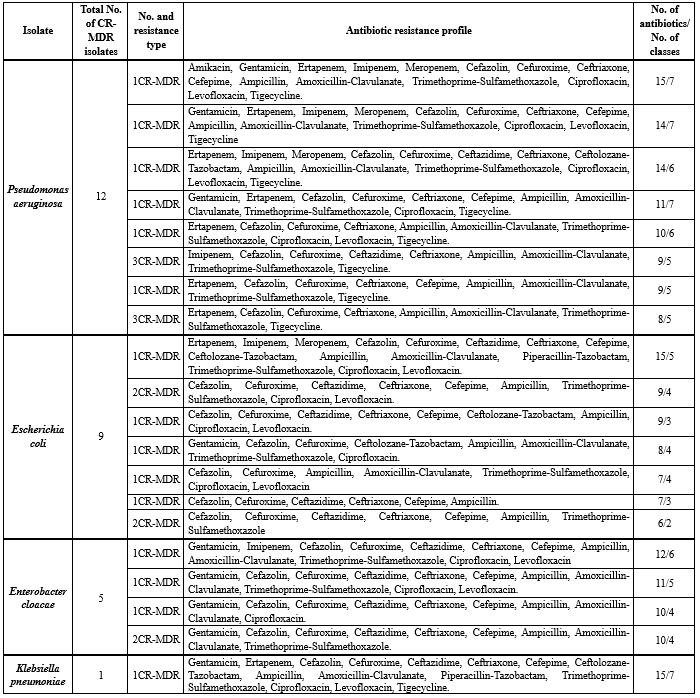 CR-MDR: Carbapenem Resistant-Multi-Drug Resistant |
Carbapenemase-producing Gram-negative isolates and their corresponding MDR patterns are detailed in Table 1. Among these isolates, 27 (38.02%) were identified as CPOs with MDR patterns. The CPO distribution showed that 12 isolates (44.44%) were carbapenemase-producing Pseudomonas aeruginosa, 9 isolates (33.33%) were carbapenemase-producing Escherichia coli, 5 isolates (18.51%) were carbapenemase-producing Enterobacter cloacae, and 1 isolate (3.7%) was carbapenemase-producing Klebsiella pneumoniae.
The antibiotic resistance profiles of carbapenem-resistant isolates revealed significant MDR patterns. Among the carbapenem-resistant Pseudomonas aeruginosa isolates, the most resistant strain demonstrated resistance to 15 antibiotics spanning 7 classes. Other P. aeruginosa isolates exhibited resistance to 14 antibiotics (7 classes), 14 antibiotics (6 classes), 11 antibiotics (7 classes), 10 antibiotics (6 classes), and multiple isolates showed resistance to either 9 antibiotics (5 classes) or 8 antibiotics (5 classes). The carbapenem-resistant Escherichia coli isolates displayed variable resistance patterns, with one isolate resistant to 15 antibiotics across 5 classes, while other isolates showed resistance ranging from 6 to 9 antibiotics covering 2 to 4 classes. Notably, the single carbapenem-resistant Klebsiella pneumoniae isolate exhibited extensive resistance to 15 antibiotics from 7 classes. The carbapenem-resistant Enterobacter cloacae isolates demonstrated resistance patterns ranging from 10 to 12 antibiotics, covering 4 to 6 different antibiotic classes, with multiple isolates showing similar resistance profiles of 10 antibiotics across 4 classes.
Discussion
CPOs have been increasingly reported worldwide among Enterobacteriaceae (9,10). However, data on carbapenem-resistant Gram-negative bacteria in Iraq remain limited. This study investigated the prevalence and resistance patterns of CPOs isolated from burn and wound infections in Iraqi hospitals.
Our findings revealed Pseudomonas aeruginosa as the most prevalent Gram-negative pathogen, consistent with its known ability to develop MDR and survive in diverse environments, leading to increased mortality and prolonged hospitalizations (11). Using the reliable RAPIDEC CARBA NP test and BD Phoenix™ M50 system, we identified 27 (38.02%) carbapenemase-producing isolates exhibiting MDR patterns. This aligns with studies from Spain, Italy, and Iraq, confirming carbapenemase production as a major resistance mechanism (12-14). Globally, CPOs pose a severe threat due to three primary resistance mechanisms: porin loss, efflux pump overexpression, and carbapenemase production. Notably, carbapenemase genes are often plasmid- or transposon-borne, facilitating horizontal gene transfer across bacterial species (15).
All studied pathogens showed high resistance to β-lactams (Penicillins and cephalosporins), likely due to β-lactamase production, altered penicillin-binding proteins, and membrane permeability changes (16). CR-P. aeruginosa and CR-K. pneumoniae exhibited the highest resistance (15 antibiotics across 7 classes). Variations in resistance patterns across studies may reflect regional differences, hospital hygiene, infection types, and detection methods (17,18). The rapid spread of MDR pathogens is exacerbated by mobile genetic elements (e.g., plasmids, transposons), which disseminate resistance genes and promote extreme drug resistance (19-21). Self-medication, antibiotic misuse, and inadequate laboratory surveillance in critical units (e.g., ICUs, burn centers) further compound this issue. As emphasized by Sadeghi et al. (22), robust surveillance programs and novel therapeutics are urgently needed to combat CPOs, which significantly increase global morbidity and mortality (23).
Conclusion
In the current study, the CPOs were highly MDR against most classes of antibiotics due to misuse or overuse of antibiotics. It is recommended that the Iraqi Ministry of Health include a new routine method in the health laboratory system for detecting the carbapenem-resistant bacteria. The wide prevalence of these organisms is in need to continuous monitoring and includes recent strategies for antibacterial resistance control and infection treatment, furthermore the discovery of new drugs.
Acknowledgement
We sincerely thank all the patients who participated in this study at the Burn Unit and Wound Care Units of Azadi Teaching Hospital in Kirkuk, Iraq, and at the Plastic, Reconstructive, and Burn Surgery Hospital in Sulaymaniyah, Kurdistan Region, Iraq. We also extend our gratitude to the dedicated medical staff at both hospitals for their invaluable support.
Funding sources
Self-funded
Ethical statement
The medical institution in Kirkuk cityIraq agreed and approved this study (Approval no. 102, approval date 5/2/2023). During this study, the research adhered to the guidelines of the Ministry of Health, Government of Iraq.
Conflicts of interest
The authors state no conflict of interest.
Author contributions
All the medical works in the current study were conducted by the author, Sarah Ahmed Hasan, and supervised by Waad Mahmood Raoof and Khaled Khalil Ahmed.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
The antibiotic resistance profiles of carbapenem-resistant isolates revealed significant MDR patterns. Among the carbapenem-resistant Pseudomonas aeruginosa isolates, the most resistant strain demonstrated resistance to 15 antibiotics spanning 7 classes. Other P. aeruginosa isolates exhibited resistance to 14 antibiotics (7 classes), 14 antibiotics (6 classes), 11 antibiotics (7 classes), 10 antibiotics (6 classes), and multiple isolates showed resistance to either 9 antibiotics (5 classes) or 8 antibiotics (5 classes). The carbapenem-resistant Escherichia coli isolates displayed variable resistance patterns, with one isolate resistant to 15 antibiotics across 5 classes, while other isolates showed resistance ranging from 6 to 9 antibiotics covering 2 to 4 classes. Notably, the single carbapenem-resistant Klebsiella pneumoniae isolate exhibited extensive resistance to 15 antibiotics from 7 classes. The carbapenem-resistant Enterobacter cloacae isolates demonstrated resistance patterns ranging from 10 to 12 antibiotics, covering 4 to 6 different antibiotic classes, with multiple isolates showing similar resistance profiles of 10 antibiotics across 4 classes.
Discussion
CPOs have been increasingly reported worldwide among Enterobacteriaceae (9,10). However, data on carbapenem-resistant Gram-negative bacteria in Iraq remain limited. This study investigated the prevalence and resistance patterns of CPOs isolated from burn and wound infections in Iraqi hospitals.
Our findings revealed Pseudomonas aeruginosa as the most prevalent Gram-negative pathogen, consistent with its known ability to develop MDR and survive in diverse environments, leading to increased mortality and prolonged hospitalizations (11). Using the reliable RAPIDEC CARBA NP test and BD Phoenix™ M50 system, we identified 27 (38.02%) carbapenemase-producing isolates exhibiting MDR patterns. This aligns with studies from Spain, Italy, and Iraq, confirming carbapenemase production as a major resistance mechanism (12-14). Globally, CPOs pose a severe threat due to three primary resistance mechanisms: porin loss, efflux pump overexpression, and carbapenemase production. Notably, carbapenemase genes are often plasmid- or transposon-borne, facilitating horizontal gene transfer across bacterial species (15).
All studied pathogens showed high resistance to β-lactams (Penicillins and cephalosporins), likely due to β-lactamase production, altered penicillin-binding proteins, and membrane permeability changes (16). CR-P. aeruginosa and CR-K. pneumoniae exhibited the highest resistance (15 antibiotics across 7 classes). Variations in resistance patterns across studies may reflect regional differences, hospital hygiene, infection types, and detection methods (17,18). The rapid spread of MDR pathogens is exacerbated by mobile genetic elements (e.g., plasmids, transposons), which disseminate resistance genes and promote extreme drug resistance (19-21). Self-medication, antibiotic misuse, and inadequate laboratory surveillance in critical units (e.g., ICUs, burn centers) further compound this issue. As emphasized by Sadeghi et al. (22), robust surveillance programs and novel therapeutics are urgently needed to combat CPOs, which significantly increase global morbidity and mortality (23).
Conclusion
In the current study, the CPOs were highly MDR against most classes of antibiotics due to misuse or overuse of antibiotics. It is recommended that the Iraqi Ministry of Health include a new routine method in the health laboratory system for detecting the carbapenem-resistant bacteria. The wide prevalence of these organisms is in need to continuous monitoring and includes recent strategies for antibacterial resistance control and infection treatment, furthermore the discovery of new drugs.
Acknowledgement
We sincerely thank all the patients who participated in this study at the Burn Unit and Wound Care Units of Azadi Teaching Hospital in Kirkuk, Iraq, and at the Plastic, Reconstructive, and Burn Surgery Hospital in Sulaymaniyah, Kurdistan Region, Iraq. We also extend our gratitude to the dedicated medical staff at both hospitals for their invaluable support.
Funding sources
Self-funded
Ethical statement
The medical institution in Kirkuk cityIraq agreed and approved this study (Approval no. 102, approval date 5/2/2023). During this study, the research adhered to the guidelines of the Ministry of Health, Government of Iraq.
Conflicts of interest
The authors state no conflict of interest.
Author contributions
All the medical works in the current study were conducted by the author, Sarah Ahmed Hasan, and supervised by Waad Mahmood Raoof and Khaled Khalil Ahmed.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Research Article: Original Paper |
Subject:
bacteriology
Received: 2024/01/17 | Accepted: 2024/04/23 | Published: 2025/09/17 | ePublished: 2025/09/17
Received: 2024/01/17 | Accepted: 2024/04/23 | Published: 2025/09/17 | ePublished: 2025/09/17
References
1. Dos Santos GS, Solidônio EG, Costa MC, Melo RO, de Souza IF, Silva G, et al. Study of the Enterobacteriaceae Group CESP (Citrobacter, Enterobacter, Serratia, Providencia, Morganella and Hafnia): A Review. The Battle Against Microbial Pathogens: Basic Science, Technological Advances and Educational Programs (A. Méndez-Vilas, Ed.) FORMATEX. 2015:794-805. [View at Publisher] [Google Scholar]
2. Patel G, Bonomo RA. Status report on carbapenemases: challenges and prospects. Expert Rev Anti Infect Ther. 2011;9(5):555-70. [View at Publisher] [DOI] [PMID] [Google Scholar]
3. Hasoon NA, Hamed SL. Molecular characterization of carbapenemase-producing Gram-negative bacteria isolated from clinical specimens in Baghdad. J Pure Appl Microbiol. 2019;13(2):1031-40. [View at Publisher] [DOI] [Google Scholar]
4. Tarashi S, Goudarzi H, Erfanimanesh S, Pormohammad A, Hashemi A. Phenotypic and molecular detection of metallo-beta-lactamase genes among imipenem resistant Pseudomonas aeruginosa and Acinetobacter baumannii strains isolated from patients with burn injuries. Arch Clin Infect Dis. 2016;11(4):e39036. [View at Publisher] [DOI] [Google Scholar]
5. Nordmann P. Carbapenemase-producing Enterobacteriaceae: overview of a major public health challenge. Med Mal Infect. 2014;44(2):51-6. [View at Publisher] [DOI] [PMID] [Google Scholar]
6. Iraz M, Düzgün AÖ, Sandallı C, Doymaz MZ, Akkoyunlu Y, Saral A, et al. Distribution of β-lactamase genes among carbapenem-resistant Klebsiella pneumoniae strains isolated from patients in Turkey. Ann Lab Med. 2015;35(6):595-601. [View at Publisher] [DOI] [PMID]
7. Bush K, Pannell M, Lock JL, Queenan AM, Jorgensen JH, Lee RM, et al. Detection systems for carbapenemase gene identification should include the SME serine carbapenemase. Int J Antimicrob Agents. 2013;41(1):1-4. [View at Publisher] [DOI] [PMID] [Google Scholar]
8. Thomson KS. Extended-spectrum-β-lactamase, AmpC, and carbapenemase issues. J Of Clin Microbiol. 2010;48(4):1019-25. [View at Publisher] [DOI] [PMID] [Google Scholar]
9. Mahrach Y, Mourabit N, Arakrak A, Bakkali M, Laglaoui A. Phenotypic and molecular study of carbapenemase-producing Enterobacteriaceae in a regional hospital in northern Morocco. J Clin Med Sci. 2019;3(1):113. [View at Publisher] [Google Scholar]
10. Hasan SA, Raoof WM, kamal Rachid S. A systematic review: The current status of carbapenem resistance in Iraq. World Bulletin of Public Health. 2022;13:88-94. [View at Publisher] [Google Scholar]
11. Hasan SA, Raoof WM, Ahmed KK. FIRST REPORT OF CO-HARBORING BLEOMYCIN RESISTANCE GENE (bleMBL) AND CARBAPENEMASE RESISTANCE GENE (blaNDM-1) KLEBSIELLA PNEUMONIAE IN IRAQ WITH COMPARISON STUDY AMONG THE SENSITIVITY TEST, THE BD PHOENIX CPO DETECT TEST, AND THE RAPIDEC® CARBA NP TEST. SJLSA. 2024;16(4):208-37. [View at Publisher] [DOI] [Google Scholar]
12. Suay-García B, Pérez-Gracia MT. Present and future of carbapenem-resistant Enterobacteriaceae infections. Advances in clinical immunology, medical microbiology, COVID-19, and big data. 2021:435-56. [View at Publisher] [DOI] [PMID] [Google Scholar]
13. Haji SH, Aka STH, Ali FA. Prevalence and characterisation of carbapenemase encoding genes in multidrug-resistant Gram-negative bacilli. PLoS One. 2021;16(11):e0259005. [View at Publisher] [DOI] [PMID] [Google Scholar]
14. Aurilio C, Sansone P, Barbarisi M, Pota V, Giaccari LG, Coppolino F, et al. Mechanisms of action of carbapenem resistance. Antibiotics. 2022;11(3):421. [View at Publisher] [DOI] [PMID] [Google Scholar]
15. Elshamy AA, Aboshanab KM. A review on bacterial resistance to carbapenems: epidemiology, detection and treatment options. Future science OA. 2020;6(3):FSO438. [View at Publisher] [DOI] [PMID] [Google Scholar]
16. Hasan SA, Abass KS. Prevalence of Gram Negative Bacteria Isolated from Patients with Burn Infection and their Antimicrobial Susceptibility Patterns in Kirkuk City, Iraq. Indian J Public Health Res Dev. 2019;10(8). [View at Publisher] [DOI] [Google Scholar]
17. Ahmed Hasan S, Raheem TF, Abdulla HM. Phenotypic, antibiotyping, and molecular detection of Klebsiella pneumoniae isolates from clinical specimens in Kirkuk, Iraq. Arch Razi Inst. 2021;76(4):1061-7. [View at Publisher] [DOI] [PMID] [Google Scholar]
18. Ahmed Hasan S, Mohammed Bakr M. Bacteriological and molecular detection of Klebsiella oxytoca and its resistance to antibiotics among clinical specimens from Kirkuk, Iraq. Arch Razi Inst. 2022;77(5):1521-5. [View at Publisher] [DOI] [PMID] [Google Scholar]
19. Workneh M, Yee R, Simner PJ. Phenotypic methods for detection of carbapenemase production in carbapenem-resistant organisms: what method should your laboratory choose?.Clin Microbiol Newsl. 2019;41(2):11-22. [View at Publisher] [DOI] [Google Scholar]
20. Hasan SA. Pseudomonas aeruginosa and the multifactorial antibiotic resistance. Eurasian Medical Research Periodical. 2022;11:85-94. [View at Publisher] [Google Scholar]
21. Fakhraddin Raheem T, Ahmed Hasan Ali S. Prevalence and multi-drug resistance patterns of uropathogenic E. coli isolated from women patients in Kirkuk City, Iraq. Iran J Microbio. 2022;16(6):609-14. [View at Publisher] [DOI] [Google Scholar]
22. Sadeghi MR, Ghotaslou R, Akhi MT, Asgharzadeh M, Hasani A. Molecular characterization of extended-spectrum β-lactamase, plasmid-mediated AmpC cephalosporinase and carbapenemase genes among Enterobacteriaceae isolates in five medical centres of East and West Azerbaijan, Iran. J Med Microbiol. 2016;65(11):1322-31. [View at Publisher] [DOI] [PMID] [Google Scholar]
23. Hasan SA, Raoof WM, Ahmed KK. Antibacterial activity of deer musk and Ziziphus spina-christi against carbapebem resis-tant gram negative bacteria isolated from patients with burns and wounds. Regulatory Mechanisms in Biosystems. 2024;15(2):267-78. [View at Publisher] [DOI] [Google Scholar]
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |








 goums.ac.ir
goums.ac.ir yahoo.com
yahoo.com