Volume 19, Issue 4 (Jul-Aug 2025)
mljgoums 2025, 19(4): 41-45 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Shareh A, Khasheii B, Faraji T, Khoshnia M, Anvari S, Jamalli A. Antibiotic resistance genes in helicobacter pylori isolates from gastric biopsies of patients with gastrointestinal diseases in Golestan province, Iran. mljgoums 2025; 19 (4) :41-45
URL: http://mlj.goums.ac.ir/article-1-1648-en.html
URL: http://mlj.goums.ac.ir/article-1-1648-en.html
Azam Shareh1 
 , Behnoush Khasheii2
, Behnoush Khasheii2 
 , Tayebeh Faraji3
, Tayebeh Faraji3 
 , Masoud Khoshnia4
, Masoud Khoshnia4 

 , Shaghayegh Anvari1
, Shaghayegh Anvari1 

 , Ailar Jamalli5
, Ailar Jamalli5 



 , Behnoush Khasheii2
, Behnoush Khasheii2 
 , Tayebeh Faraji3
, Tayebeh Faraji3 
 , Masoud Khoshnia4
, Masoud Khoshnia4 

 , Shaghayegh Anvari1
, Shaghayegh Anvari1 

 , Ailar Jamalli5
, Ailar Jamalli5 


1- Laboratory Sciences Research Center, Golestan University of Medical Sciences, Gorgan, Iran
2- Department of Pathobiology, Faculty of Veterinary Sciences, Bu-Ali Sina University, Hamedan, Iran
3- Department of Microbiology, Faculty of Medical Sciences, Golestan University of Medical Sciences, Gorgan, Iran
4- Golestan Research Center of Gastroenterology and Hepatology, Golestan University of Medical Sciences, Gorgan, Iran
5- Laboratory Sciences Research Center, Golestan University of Medical Sciences, Gorgan, Iran ,jamali@goums.ac.ir
2- Department of Pathobiology, Faculty of Veterinary Sciences, Bu-Ali Sina University, Hamedan, Iran
3- Department of Microbiology, Faculty of Medical Sciences, Golestan University of Medical Sciences, Gorgan, Iran
4- Golestan Research Center of Gastroenterology and Hepatology, Golestan University of Medical Sciences, Gorgan, Iran
5- Laboratory Sciences Research Center, Golestan University of Medical Sciences, Gorgan, Iran ,
Keywords: Helicobacter pylori, Drug resistance, Microbial, Polymerase Chain Reaction, Polymorphism, Restriction Fragment Length
Full-Text [PDF 457 kb]
(794 Downloads)
| Abstract (HTML) (3118 Views)
Gene sequencing for each sample was performed using Chromas software (Version 2.6.4) for the genes of H. pylori 26695 at https://blast.ncbi.nlm.nih.gov/. The sequences were then compared and analyzed using BLAST. Detection of point mutations in the second V of 23S rRNA gene related to clarithromycin resistance of H. pylori was performed by the PCR-RLFP method. Amplification was carried out using the polymerase chain reaction (PCR) technique (Table 1) (21).
The PCR procedure details in this study were as follows. To prepare the PCR reaction, 2.5 μL of PCR master mix (1×) was combined with 1 μL of each primer, 5 μL of template DNA, and 13.1 μL of distilled water. The master mix contained Taq DNA polymerase (0.08 U/μL), MgCl2 (1.5 mM), and dNTPs (0.2 mM). We used BsaI and BbsI (Thermo) enzymes to detect A2143G and A2142G point mutations, respectively.
The PCR-RFLP protocol for detecting mutations at the A2143G locus using the BsaI enzyme was as follows. 10 µL of PCR products, along with 2 µL of 10X buffer G, 1 µL of BsaI enzyme, and 16 µL of sterile distilled water, were added to microtubes. The PCR-RFLP protocol for detecting mutations at the A2142G site using the BbsI enzyme was as follows. 10 µL of PCR products with 2 µl of 10X buffer G, 1 µl of BbsI enzyme, and 18 µL of sterile distilled water were mixed in a microtube. The microtubules were then spun for a few seconds and then incubated at 37 °C for three hours. To inactivate the enzyme, the sample was incubated for 20 minutes at 65 °C. Ultimately, the digested PCR and undigested PCR products were examined by UV transilluminator after electrophoresis in 2% agarose gel stained with ethidium bromide.
The data were analyzed using SPSS 16 software. P values were obtained through the Chi-square test, with P values < 0.05 considered statistically significant in all cases.
Results
In total, 335 biopsy specimens were collected from patients over the course of one year, from July 2016 to July 2017. Out of all 335 specimens, 37.9% (127) were positive for urease isolates. Among 127 urease-positive isolates (81.88%), 80 had a conserved ureC gene, including 54 females (67.5%) and 26 males (32.5%).
Prevalence of clarithromycin resistance
After amplifying the target fragment by PCR using specific primers for the 23S rRNA gene and electrophoresis of PCR products, a 425 bp band was created. We examined clarithromycin resistance and mutations at the A2142G and A2143G loci using BbsI and BsaI enzymes, respectively. The BsaI enzyme has two cleavage sites on the PCR product, 425 bp. Three fragments were created by enzymatic digestion of DNA with lengths of 20 bp, 101 bp, and 304 bp. The 20bp fragment is not visible on the gel (Figure 1). The BbsI enzyme created a cleavage site through the enzymatic digestion of two pieces, 93 bp and 332 bp. Among the 80 isolates, 25% were resistant to clarithromycin; 35% had the A2142G mutation, and 65% had the A2143G mutation.
Our study found no deletion of 200 nucleotides in the studied isolates that confer metronidazole resistance.
Prevalence of fluoroquinolone resistance
Out of the 80 isolates studied, 22 isolates (27.5%) exhibited resistance to fluoroquinolones. Among these resistant isolates, there were 8 mutations (36.36%) at the 87th amino acid position, where asparagine is located, and 14 mutations (63.63%) at the 91st amino acid position, where aspartate is found. At position 87, the highest mutation leading to resistance to fluoroquinolones was due to a change in the amino acid asparagine to lysine (31.82%). Another mutation reported in some studies is a change in the amino acid asparagine to isoleucine (4.54%), which was observed in one of the isolates. At position 91, changes in the amino acid aspartate to glycine (27.27%), aspartate to asparagine (22.72%), and aspartate to tyrosine (13.63%) were observed among the isolates in this study, and two isolates (2.06%) had a D91A mutation. Among the specimens studies, these mutations did not lead to resistance to fluoroquinolones.
The highest antibiotic resistance among the isolates was against fluoroquinolone (27.5%), and metronidazole resistance was the lowest. None of the 80 samples had 200 nucleotide deletions. Concomitant resistance to clarithromycin and fluoroquinolone was observed in 11 samples (13.75%) of H. pylori. Additionally, no statistically significant association was found between antibiotic resistance and age, sex, or patients' symptoms, including a history of peptic ulcer and gastrointestinal problems.
Discussion
Antibiotic resistance in H. pylori is a crucial factor influencing treatment regimens (22). Various antibiotics are used to treat H. pylori infection, including metronidazole, clarithromycin, levofloxacin, and amoxicillin (23). This study investigated the antibiotic resistance of H. pylori strains and resistance-causing mutations by PCR and PCR-RFLP.
Metronidazole resistance is a global concern (24). From 1997 to 2013, epidemiological studies in Iran reported the highest antibiotic resistance of H. pylori to metronidazole, with a rate of 61.6% (25). Abdollahi et al. (2010) in Iran conducted a study on 63 isolates of H. pylori, of which 35 samples were resistant to metronidazole. Examining the rdxA gene demonstrated that eight isolates (22.9%) had 200 nucleotide deletions (26). Ramzy et al. examined the rdxA gene in Egypt in 2013 and observed the removal of 200 nucleotides from 70 samples in 44 strains (62.9%) (27). In a 2013 study by Mirzaei et al. in Iran, 27 of 48 patients had metronidazole resistance. They were tested for the rdxA gene, and no deletion of 200 nucleotides was observed (28). Mahmoudi et al. (2015) conducted a study in Iran on 46 H. pylori isolates. The findings of this study indicated that 64.3% of the isolates were resistant to metronidazole. Notably, none of the resistant samples demonstrated deletion of the rdxA gene (20).
Our study confirmed the findings of Mirzaei and Mahmoudi; none of the strains we studied exhibited the removal of 200 nucleotides. The diversity of clarithromycin resistance in different regions highlights the need to study the level of resistance in each geographical area to inform more effective treatment regimens (29). Clarithromycin is a macrolide (30), and the most important related mutations are 2142 and 2143 (10). Clarithromycin resistance rates vary across different regions in Iran, with the lowest rate observed in Rasht at 5.5% and the highest rate in Kashan at 33.7% (31). Clarithromycin resistance can arise from the A2143G mutation, which occurs with a frequency of 69.8%, the A2142G mutation with a frequency of 11.7%, and the A2142C mutation with a frequency of 2.6% (32-34). Sadeghifard et al. conducted a study in 2010 on 50 patients to evaluate clarithromycin resistance in Iran. Eight patients (16%) had clarithromycin resistance at the A2143G locus, and no mutation was observed at the A2142G locus (35). In 2011, Abdollahi et al. conducted a study in the same country on 63 H. pylori isolates obtained from gastric biopsy specimens to investigate the resistance and sensitivity of the isolates to clarithromycin and to investigate mutations in the 23S RRNA gene. They found that 20 out of 63 H. pylori isolates (31.7%) were resistant to clarithromycin. Among these resistant isolates, 3 (15%) exhibited mutations at the A2143G locus, 11 (55%) at the A2142G locus, and 6 (30%) at the A2142C locus (36). Also, Eghbali et al. in 2016 in Iran conducted a study on 89 gastric biopsies of patients to investigate the resistance of H. pylori strains to clarithromycin; 5 strains were resistant to clarithromycin and had A2143G mutation, and no strains had A2142G mutation (37). Suzuki et al. investigated clarithromycin resistance in 488 H. pylori cases in 2013 in Brazil and observed 25% A2142G mutations, 58.3% A2143G mutations, and 8.7% mutations at both sites (38). In a 2020 study by Vazirzadeh et al. in Iran, 21 (25.3%) out of 83 H. pylori strains were resistant to clarithromycin, and 19 isolates had mutations. Among them, 13 strains (68.4%) had A2143G mutation and 6 strains (31.5%) had A2144G mutation; the mutation A2143C was not detected in any of the isolated (39).
Our findings indicated that the resistance to clarithromycin was 25%. The mutation at the A2143G site was 65%, and the mutation at the A2142G site was 35%, similar to the Suzuki and Vazirizadeh studies. The mechanism of action of fluoroquinolones is through their effect on H.pylori DNA gyrase (40). In 2015, Zaki et al. in Egypt investigated mutations in the gyrA gene at positions 87 and 91 on 82 samples. Nineteen strains had mutations in gyrA, of which 12 isolates (63.2%) had mutations in aspartate 91, and 7 isolates (36.8%) had mutations in asparagine 87 (41).
In 2014, Ngoyi et al. conducted a study in the Congo on the resistance of 36 patient samples to fluoroquinolones. They found that 18 (50%) of the H. pylori isolates had mutations at positions 87 and 91 (42). In 2020, Kipritci et al. conducted a study on 140 biopsy specimens in Turkey to investigate the antibiotic resistance of H. pylori strains to clarithromycin and fluoroquinolones. The results indicated that 21 isolates (25.6%) showed resistance to fluoroquinolones, while 31 isolates (37.9%) exhibited resistance to clarithromycin. Additionally, 11 isolates (14.1%) demonstrated simultaneous resistance to fluoroquinolones and clarithromycin (43). In this study, the resistance to fluoroquinolones was 27.5%, which was 63% in the 91-amino acid position of aspartate and 36% in the 87-amino acid position of asparagine. Our study was similar to the Zaki study. Additionally, identical to the Kipritci study, our study demonstrated simultaneous resistance to fluoroquinolones and clarithromycin. Resistance to fluoroquinolones seems to be due to mutations in the gyrA gene. Increased resistance to fluoroquinolones may be associated with excessive use of these antibiotics. Since this antibiotic has a wide range of therapeutic applications and is used against various types of infections, it is not recommended for the treatment of H. pylori. The resistance rate to fluoroquinolones in Iran increased from 5.3% in 1997 to 27.5% in 2013.
Conclusion
Antibiotic resistance among H. pylori strains is rising globally, leading to treatment failures. Since resistance to metronidazole, clarithromycin, and fluoroquinolones has become a grave concern, alternative treatment regimens should be considered if resistance to clarithromycin and fluoroquinolones is suspected. Considering high levels of metronidazole resistance worldwide, metronidazole resistance should be further investigated using phenotypic methods and PCR techniques. It is also recommended to study other genes involved in metronidazole activation, such as frxA, fdxA, fdxB. To investigate clarithromycin resistance, research should not be limited to A2143G and A2142G mutations, which are the most common mutations associated with resistance, and other mutations should also be considered.
Acknowledgement
We extend our gratitude to the staff of Golestan University of Medical Sciences, Naemeh Javid, and Masoud Bazori, for their logistical support. This work was funded by the Technology and Research Deputy of Golestan University of Medical Sciences (Contract no. 9412183500).
Funding sources
This research was funded by the Technology and Research Deputy of Golestan University of Medical Sciences.
Ethical statement
Informed consent was obtained from all participants. The study received approval from the Human Research Ethics Boards at Golestan University of Medical Sciences under the Code of Ethics (IR.GOUMS.REC.1394.351).
Conflicts of interest
The authors declare that there are no conflicts of interest.
Author contributions
A.SH conducted the experiments and analyzed the data. B. KH wrote the manuscript and contributed to the article's concept. A. J and SH guided the experiments and critically revised the manuscript.
Data availability statement
All data generated and analyzed during this study are included in our manuscript.
Full-Text: (646 Views)
Introduction
Helicobacter pylori (H. pylori) is a gram-negative, flagellated, microaerophilic bacterium (1). H. pylori infection is a major cause of gastrointestinal diseases, including chronic gastritis, peptic ulcer, and gastric cancer (2). The prevalence of this infection in Iran is 60 to 90%, which indicates that Iran is a high-risk area for H. pylori infection (3,4). Virulence factors in the pathogenesis of H. pylori include urease, flagellum, binding, cytotoxin-associated gene A (cagA), and vacuolating cytotoxin A (vacA) (5). The prevalence of H. pylori antibiotic resistance varies from country to country and may be related to geographical factors (6).
Treatment for H. pylori often involves the use of antibiotics. A common reason for treatment failure is the bacteria's ability to colonize the subcutaneous layer, and when it is combined with stomach acid, it can reduce the effectiveness of these antibiotics (6,7). A PPI (Proton pump inhibitor) or ranitidine, bismuth, and two antibiotics, such as amoxicillin and clarithromycin, are often used for its treatment (8). While three-drug regimens typically succeed in eradicating H. pylori infections, rising resistance-particularly to clarithromycin and metronidazole-among clinical strains can lead to treatment failures (9). Clarithromycin is a macrolide antibiotic that inhibits protein synthesis by binding to the 23S rRNA peptidyl transferase region. Clarithromycin resistance in H. pylori results from specific point mutations in the peptidyl transferase region of the second V23S rRNA gene (10). The most common mutation is the replacement of A to G at positions 2143 (A2143G) and 2142 (A2142G) (11,12). Metronidazole is a prodrug that requires regeneration for activation.
Several nitroreductases in H. pylori regenerate nitroimidazoles. Reduction of the nitro group produces imidazole mediators and toxic radicals that are toxic to DNA. One of the essential nitroreductases is oxygen-insensitive NAD(P)H, which is encoded by the rdxA gene; a mutation in this gene can lead to resistance to metronidazole (13). Fluoroquinolones, which inhibit the growth of H. pylori in vitro (14), can affect DNA gyrase (15). The resistance mechanism to fluoroquinolones in H. pylori is related to mutations in the regions determining the quinolone resistance of gyrA (QRDRs) and is caused by point mutations in the gyrA gene encoding gyrase DNA at positions encoding amino acids N87, 88A, 91D, and 97A (16). This study aimed to determine the antibiotic resistance of Helicobacter pylori strains in biopsy specimens and to investigate mutations in genes that confer resistance to clarithromycin, metronidazole, and fluoroquinolones using PCR and PCR-RFLP methods in specimens collected from Golestan Province, Iran.
Methods
In this study, 80 biopsy specimens from 335 patients referred to Sayad Shirazi Hospital in Gorgan for gastrointestinal issues and gastric ulcers were analyzed. The specimens were collected over a period of one year, from July 2016 to July 2017, and the patients' ages ranged from 14 to 86 years. The gastric biopsy specimens were taken during endoscopy by a gastroenterologist with a sterile endoscope.
Two gastric biopsies were taken from each patient. One sample was transferred to the urease medium for a rapid urease test (RUT) for H. pylori screening, and a second sample was transferred to 1.5 mL microtubes containing 1 mL of sterile phosphate-buffered saline (PBS) until DNA extraction. For Subsequent steps, the samples were kept at -70 °C. Informed consent was obtained from the patient before participating in this study. The study was approved by Golestan University of Medical Sciences, Iran. Ethical approval was granted under the Code of Ethics (IR.GOUMS.REC.1394.351).
The RUT was conducted following the method outlined by Siavoshi et al (17). Aliquots were collected in sterilized microtubules with a 700-1000 microliter volume. One of the biopsy specimens was placed in a urease medium for 1-2 hours at room temperature, and the change in color from yellow to pink indicated a positive test. Samples of patients whose urease test was positive were included in the study and kept at -70. According to the kit manufacturer's protocol, the total genomic DNA of 80 H. pylori strains was extracted using the Macherey-Nagel Kit (Germany). The extracted DNAs were stored for further analysis at -20° C. All primers for PCR in this study were synthesized by the German Metabion.
The primers described by Momtaz et al (18) were used to identify the ureC gene (Table 1). PCR amplification was performed to investigate the resistance of H. pylori isolates to fluoroquinolones (gyrA) and metronidazole (rdxA). The primers developed by Glocker et al. and Mahmoudi et al. were used to identify the gyrA and rdxA genes (19,20) (Table 1). To identify the gyrA and ureC genes, a PCR master mix was prepared by combining 2.5 μL of 1× PCR master mix, which contained Taq DNA polymerase (0.06 U/μL), MgCl2 (1.5 mM), and dNTPs (0.2 mM). This was mixed with 1 μL of each primer, 5 μL of template DNA, and 13.2 μL of distilled water.
Helicobacter pylori (H. pylori) is a gram-negative, flagellated, microaerophilic bacterium (1). H. pylori infection is a major cause of gastrointestinal diseases, including chronic gastritis, peptic ulcer, and gastric cancer (2). The prevalence of this infection in Iran is 60 to 90%, which indicates that Iran is a high-risk area for H. pylori infection (3,4). Virulence factors in the pathogenesis of H. pylori include urease, flagellum, binding, cytotoxin-associated gene A (cagA), and vacuolating cytotoxin A (vacA) (5). The prevalence of H. pylori antibiotic resistance varies from country to country and may be related to geographical factors (6).
Treatment for H. pylori often involves the use of antibiotics. A common reason for treatment failure is the bacteria's ability to colonize the subcutaneous layer, and when it is combined with stomach acid, it can reduce the effectiveness of these antibiotics (6,7). A PPI (Proton pump inhibitor) or ranitidine, bismuth, and two antibiotics, such as amoxicillin and clarithromycin, are often used for its treatment (8). While three-drug regimens typically succeed in eradicating H. pylori infections, rising resistance-particularly to clarithromycin and metronidazole-among clinical strains can lead to treatment failures (9). Clarithromycin is a macrolide antibiotic that inhibits protein synthesis by binding to the 23S rRNA peptidyl transferase region. Clarithromycin resistance in H. pylori results from specific point mutations in the peptidyl transferase region of the second V23S rRNA gene (10). The most common mutation is the replacement of A to G at positions 2143 (A2143G) and 2142 (A2142G) (11,12). Metronidazole is a prodrug that requires regeneration for activation.
Several nitroreductases in H. pylori regenerate nitroimidazoles. Reduction of the nitro group produces imidazole mediators and toxic radicals that are toxic to DNA. One of the essential nitroreductases is oxygen-insensitive NAD(P)H, which is encoded by the rdxA gene; a mutation in this gene can lead to resistance to metronidazole (13). Fluoroquinolones, which inhibit the growth of H. pylori in vitro (14), can affect DNA gyrase (15). The resistance mechanism to fluoroquinolones in H. pylori is related to mutations in the regions determining the quinolone resistance of gyrA (QRDRs) and is caused by point mutations in the gyrA gene encoding gyrase DNA at positions encoding amino acids N87, 88A, 91D, and 97A (16). This study aimed to determine the antibiotic resistance of Helicobacter pylori strains in biopsy specimens and to investigate mutations in genes that confer resistance to clarithromycin, metronidazole, and fluoroquinolones using PCR and PCR-RFLP methods in specimens collected from Golestan Province, Iran.
Methods
In this study, 80 biopsy specimens from 335 patients referred to Sayad Shirazi Hospital in Gorgan for gastrointestinal issues and gastric ulcers were analyzed. The specimens were collected over a period of one year, from July 2016 to July 2017, and the patients' ages ranged from 14 to 86 years. The gastric biopsy specimens were taken during endoscopy by a gastroenterologist with a sterile endoscope.
Two gastric biopsies were taken from each patient. One sample was transferred to the urease medium for a rapid urease test (RUT) for H. pylori screening, and a second sample was transferred to 1.5 mL microtubes containing 1 mL of sterile phosphate-buffered saline (PBS) until DNA extraction. For Subsequent steps, the samples were kept at -70 °C. Informed consent was obtained from the patient before participating in this study. The study was approved by Golestan University of Medical Sciences, Iran. Ethical approval was granted under the Code of Ethics (IR.GOUMS.REC.1394.351).
The RUT was conducted following the method outlined by Siavoshi et al (17). Aliquots were collected in sterilized microtubules with a 700-1000 microliter volume. One of the biopsy specimens was placed in a urease medium for 1-2 hours at room temperature, and the change in color from yellow to pink indicated a positive test. Samples of patients whose urease test was positive were included in the study and kept at -70. According to the kit manufacturer's protocol, the total genomic DNA of 80 H. pylori strains was extracted using the Macherey-Nagel Kit (Germany). The extracted DNAs were stored for further analysis at -20° C. All primers for PCR in this study were synthesized by the German Metabion.
The primers described by Momtaz et al (18) were used to identify the ureC gene (Table 1). PCR amplification was performed to investigate the resistance of H. pylori isolates to fluoroquinolones (gyrA) and metronidazole (rdxA). The primers developed by Glocker et al. and Mahmoudi et al. were used to identify the gyrA and rdxA genes (19,20) (Table 1). To identify the gyrA and ureC genes, a PCR master mix was prepared by combining 2.5 μL of 1× PCR master mix, which contained Taq DNA polymerase (0.06 U/μL), MgCl2 (1.5 mM), and dNTPs (0.2 mM). This was mixed with 1 μL of each primer, 5 μL of template DNA, and 13.2 μL of distilled water.
|
Table 1. Primer pairs for the PCR amplification of ureC, gyrA, rdxA, and 23S rRNA genes in H. pylori isolates
 |
Gene sequencing for each sample was performed using Chromas software (Version 2.6.4) for the genes of H. pylori 26695 at https://blast.ncbi.nlm.nih.gov/. The sequences were then compared and analyzed using BLAST. Detection of point mutations in the second V of 23S rRNA gene related to clarithromycin resistance of H. pylori was performed by the PCR-RLFP method. Amplification was carried out using the polymerase chain reaction (PCR) technique (Table 1) (21).
The PCR procedure details in this study were as follows. To prepare the PCR reaction, 2.5 μL of PCR master mix (1×) was combined with 1 μL of each primer, 5 μL of template DNA, and 13.1 μL of distilled water. The master mix contained Taq DNA polymerase (0.08 U/μL), MgCl2 (1.5 mM), and dNTPs (0.2 mM). We used BsaI and BbsI (Thermo) enzymes to detect A2143G and A2142G point mutations, respectively.
The PCR-RFLP protocol for detecting mutations at the A2143G locus using the BsaI enzyme was as follows. 10 µL of PCR products, along with 2 µL of 10X buffer G, 1 µL of BsaI enzyme, and 16 µL of sterile distilled water, were added to microtubes. The PCR-RFLP protocol for detecting mutations at the A2142G site using the BbsI enzyme was as follows. 10 µL of PCR products with 2 µl of 10X buffer G, 1 µl of BbsI enzyme, and 18 µL of sterile distilled water were mixed in a microtube. The microtubules were then spun for a few seconds and then incubated at 37 °C for three hours. To inactivate the enzyme, the sample was incubated for 20 minutes at 65 °C. Ultimately, the digested PCR and undigested PCR products were examined by UV transilluminator after electrophoresis in 2% agarose gel stained with ethidium bromide.
The data were analyzed using SPSS 16 software. P values were obtained through the Chi-square test, with P values < 0.05 considered statistically significant in all cases.
Results
In total, 335 biopsy specimens were collected from patients over the course of one year, from July 2016 to July 2017. Out of all 335 specimens, 37.9% (127) were positive for urease isolates. Among 127 urease-positive isolates (81.88%), 80 had a conserved ureC gene, including 54 females (67.5%) and 26 males (32.5%).
Prevalence of clarithromycin resistance
After amplifying the target fragment by PCR using specific primers for the 23S rRNA gene and electrophoresis of PCR products, a 425 bp band was created. We examined clarithromycin resistance and mutations at the A2142G and A2143G loci using BbsI and BsaI enzymes, respectively. The BsaI enzyme has two cleavage sites on the PCR product, 425 bp. Three fragments were created by enzymatic digestion of DNA with lengths of 20 bp, 101 bp, and 304 bp. The 20bp fragment is not visible on the gel (Figure 1). The BbsI enzyme created a cleavage site through the enzymatic digestion of two pieces, 93 bp and 332 bp. Among the 80 isolates, 25% were resistant to clarithromycin; 35% had the A2142G mutation, and 65% had the A2143G mutation.
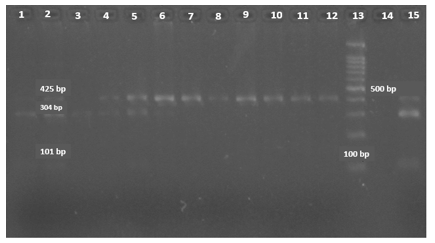 Figure 1. PCR-RFLP analysis of a 425 bp fragment of the 23S rRNA gene; Samples No. 1 to 6 and 15 are clarithromycin-resistant strains using the BsaI enzyme with A2143G mutation; Samples No. 7 to 12 are clarithromycin-sensitive strains; Sample No. 13 is a 100 bp DNA marker; Sample No. 14 is without a sample. |
Our study found no deletion of 200 nucleotides in the studied isolates that confer metronidazole resistance.
Prevalence of fluoroquinolone resistance
Out of the 80 isolates studied, 22 isolates (27.5%) exhibited resistance to fluoroquinolones. Among these resistant isolates, there were 8 mutations (36.36%) at the 87th amino acid position, where asparagine is located, and 14 mutations (63.63%) at the 91st amino acid position, where aspartate is found. At position 87, the highest mutation leading to resistance to fluoroquinolones was due to a change in the amino acid asparagine to lysine (31.82%). Another mutation reported in some studies is a change in the amino acid asparagine to isoleucine (4.54%), which was observed in one of the isolates. At position 91, changes in the amino acid aspartate to glycine (27.27%), aspartate to asparagine (22.72%), and aspartate to tyrosine (13.63%) were observed among the isolates in this study, and two isolates (2.06%) had a D91A mutation. Among the specimens studies, these mutations did not lead to resistance to fluoroquinolones.
The highest antibiotic resistance among the isolates was against fluoroquinolone (27.5%), and metronidazole resistance was the lowest. None of the 80 samples had 200 nucleotide deletions. Concomitant resistance to clarithromycin and fluoroquinolone was observed in 11 samples (13.75%) of H. pylori. Additionally, no statistically significant association was found between antibiotic resistance and age, sex, or patients' symptoms, including a history of peptic ulcer and gastrointestinal problems.
Discussion
Antibiotic resistance in H. pylori is a crucial factor influencing treatment regimens (22). Various antibiotics are used to treat H. pylori infection, including metronidazole, clarithromycin, levofloxacin, and amoxicillin (23). This study investigated the antibiotic resistance of H. pylori strains and resistance-causing mutations by PCR and PCR-RFLP.
Metronidazole resistance is a global concern (24). From 1997 to 2013, epidemiological studies in Iran reported the highest antibiotic resistance of H. pylori to metronidazole, with a rate of 61.6% (25). Abdollahi et al. (2010) in Iran conducted a study on 63 isolates of H. pylori, of which 35 samples were resistant to metronidazole. Examining the rdxA gene demonstrated that eight isolates (22.9%) had 200 nucleotide deletions (26). Ramzy et al. examined the rdxA gene in Egypt in 2013 and observed the removal of 200 nucleotides from 70 samples in 44 strains (62.9%) (27). In a 2013 study by Mirzaei et al. in Iran, 27 of 48 patients had metronidazole resistance. They were tested for the rdxA gene, and no deletion of 200 nucleotides was observed (28). Mahmoudi et al. (2015) conducted a study in Iran on 46 H. pylori isolates. The findings of this study indicated that 64.3% of the isolates were resistant to metronidazole. Notably, none of the resistant samples demonstrated deletion of the rdxA gene (20).
Our study confirmed the findings of Mirzaei and Mahmoudi; none of the strains we studied exhibited the removal of 200 nucleotides. The diversity of clarithromycin resistance in different regions highlights the need to study the level of resistance in each geographical area to inform more effective treatment regimens (29). Clarithromycin is a macrolide (30), and the most important related mutations are 2142 and 2143 (10). Clarithromycin resistance rates vary across different regions in Iran, with the lowest rate observed in Rasht at 5.5% and the highest rate in Kashan at 33.7% (31). Clarithromycin resistance can arise from the A2143G mutation, which occurs with a frequency of 69.8%, the A2142G mutation with a frequency of 11.7%, and the A2142C mutation with a frequency of 2.6% (32-34). Sadeghifard et al. conducted a study in 2010 on 50 patients to evaluate clarithromycin resistance in Iran. Eight patients (16%) had clarithromycin resistance at the A2143G locus, and no mutation was observed at the A2142G locus (35). In 2011, Abdollahi et al. conducted a study in the same country on 63 H. pylori isolates obtained from gastric biopsy specimens to investigate the resistance and sensitivity of the isolates to clarithromycin and to investigate mutations in the 23S RRNA gene. They found that 20 out of 63 H. pylori isolates (31.7%) were resistant to clarithromycin. Among these resistant isolates, 3 (15%) exhibited mutations at the A2143G locus, 11 (55%) at the A2142G locus, and 6 (30%) at the A2142C locus (36). Also, Eghbali et al. in 2016 in Iran conducted a study on 89 gastric biopsies of patients to investigate the resistance of H. pylori strains to clarithromycin; 5 strains were resistant to clarithromycin and had A2143G mutation, and no strains had A2142G mutation (37). Suzuki et al. investigated clarithromycin resistance in 488 H. pylori cases in 2013 in Brazil and observed 25% A2142G mutations, 58.3% A2143G mutations, and 8.7% mutations at both sites (38). In a 2020 study by Vazirzadeh et al. in Iran, 21 (25.3%) out of 83 H. pylori strains were resistant to clarithromycin, and 19 isolates had mutations. Among them, 13 strains (68.4%) had A2143G mutation and 6 strains (31.5%) had A2144G mutation; the mutation A2143C was not detected in any of the isolated (39).
Our findings indicated that the resistance to clarithromycin was 25%. The mutation at the A2143G site was 65%, and the mutation at the A2142G site was 35%, similar to the Suzuki and Vazirizadeh studies. The mechanism of action of fluoroquinolones is through their effect on H.pylori DNA gyrase (40). In 2015, Zaki et al. in Egypt investigated mutations in the gyrA gene at positions 87 and 91 on 82 samples. Nineteen strains had mutations in gyrA, of which 12 isolates (63.2%) had mutations in aspartate 91, and 7 isolates (36.8%) had mutations in asparagine 87 (41).
In 2014, Ngoyi et al. conducted a study in the Congo on the resistance of 36 patient samples to fluoroquinolones. They found that 18 (50%) of the H. pylori isolates had mutations at positions 87 and 91 (42). In 2020, Kipritci et al. conducted a study on 140 biopsy specimens in Turkey to investigate the antibiotic resistance of H. pylori strains to clarithromycin and fluoroquinolones. The results indicated that 21 isolates (25.6%) showed resistance to fluoroquinolones, while 31 isolates (37.9%) exhibited resistance to clarithromycin. Additionally, 11 isolates (14.1%) demonstrated simultaneous resistance to fluoroquinolones and clarithromycin (43). In this study, the resistance to fluoroquinolones was 27.5%, which was 63% in the 91-amino acid position of aspartate and 36% in the 87-amino acid position of asparagine. Our study was similar to the Zaki study. Additionally, identical to the Kipritci study, our study demonstrated simultaneous resistance to fluoroquinolones and clarithromycin. Resistance to fluoroquinolones seems to be due to mutations in the gyrA gene. Increased resistance to fluoroquinolones may be associated with excessive use of these antibiotics. Since this antibiotic has a wide range of therapeutic applications and is used against various types of infections, it is not recommended for the treatment of H. pylori. The resistance rate to fluoroquinolones in Iran increased from 5.3% in 1997 to 27.5% in 2013.
Conclusion
Antibiotic resistance among H. pylori strains is rising globally, leading to treatment failures. Since resistance to metronidazole, clarithromycin, and fluoroquinolones has become a grave concern, alternative treatment regimens should be considered if resistance to clarithromycin and fluoroquinolones is suspected. Considering high levels of metronidazole resistance worldwide, metronidazole resistance should be further investigated using phenotypic methods and PCR techniques. It is also recommended to study other genes involved in metronidazole activation, such as frxA, fdxA, fdxB. To investigate clarithromycin resistance, research should not be limited to A2143G and A2142G mutations, which are the most common mutations associated with resistance, and other mutations should also be considered.
Acknowledgement
We extend our gratitude to the staff of Golestan University of Medical Sciences, Naemeh Javid, and Masoud Bazori, for their logistical support. This work was funded by the Technology and Research Deputy of Golestan University of Medical Sciences (Contract no. 9412183500).
Funding sources
This research was funded by the Technology and Research Deputy of Golestan University of Medical Sciences.
Ethical statement
Informed consent was obtained from all participants. The study received approval from the Human Research Ethics Boards at Golestan University of Medical Sciences under the Code of Ethics (IR.GOUMS.REC.1394.351).
Conflicts of interest
The authors declare that there are no conflicts of interest.
Author contributions
A.SH conducted the experiments and analyzed the data. B. KH wrote the manuscript and contributed to the article's concept. A. J and SH guided the experiments and critically revised the manuscript.
Data availability statement
All data generated and analyzed during this study are included in our manuscript.
Research Article: Research Article |
Subject:
Microbiology
Received: 2023/03/25 | Accepted: 2023/07/9 | Published: 2025/08/31 | ePublished: 2025/08/31
Received: 2023/03/25 | Accepted: 2023/07/9 | Published: 2025/08/31 | ePublished: 2025/08/31
References
1. Wu W, Yang Y, Sun G. Recent insights into antibiotic resistance in Helicobacter pylori eradication. Gastroenterol Res Pract. 2012;2012:723183. [View at Publisher] [DOI] [PMID] [Google Scholar]
2. Ishaq S, Nunn L. Helicobacter pylori and gastric cancer: a state of the art review. Gastroenterol Hepatol Bed Bench .2015;8(Suppl1):S6-S14. [View at Publisher] [DOI] [PMID] [Google Scholar]
3. Dabiri H, Maleknejad P, Yamaoka Y, Feizabadi MM, Jafari F, Rezadehbashi M, et al. Distribution of Helicobacter pylori cagA, cagE, oipA and vacA in different major ethnic groups in Tehran, Iran. J Gastroenterol Hepatol. 2009;24(8):1380-6. [View at Publisher] [DOI] [PMID] [Google Scholar]
4. Jafari F, Shokrzadeh L, Dabiri H, Baghaei K, Yamaoka Y, Zojaji H, et al. vacA genotypes of Helicobacter pylori in relation to cagA status and clinical outcomes in Iranian populations. Jpn J Infect Dis. 2008;61(4):290-3. [View at Publisher] [DOI] [PMID] [Google Scholar]
5. Chang W-L, Yeh Y-C, Sheu B-S. The impacts of H. pylori virulence factors on the development of gastroduodenal diseases. J Biomed Sci. 2018;25(1):1-9. [View at Publisher] [DOI] [PMID] [Google Scholar]
6. De Francesco V, Giorgio F, Hassan C, Manes G, Vannella L, Panella C, et al. Worldwide H. pylori antibiotic resistance: a systematic review. J Gastrointestin Liver Dis. 2010;19(4):409-14. [View at Publisher] [PMID] [Google Scholar]
7. Nishizawa T, Suzuki H. Mechanisms of Helicobacter pylori antibiotic resistance and molecular testing. Front Mol Biosci. 2014;1:19. [View at Publisher] [DOI] [PMID] [Google Scholar]
8. Vale FF, Rosa MR, Oleastro M. Helicobacter pylori resistance to antibiotics. Science against microbial pathogens: communicating current research and technological advances. 2011:p745-56. [View at Publisher] [Google Scholar]
9. Karczewska E, Wojtas-Bonior I, Sito E, Zwolińska-Wcisło M, Budak A. Primary and secondary clarithromycin, metronidazole, amoxicillin and levofloxacin resistance to Helicobacter pylori in southern Poland. Pharmacol Rep. 2011;63(3):799-807. [View at Publisher] [DOI] [PMID] [Google Scholar]
10. Agudo S, Pérez-Pérez G, Alarcón T, López-Brea M. High prevalence of clarithromycin-resistant Helicobacter pylori strains and risk factors associated with resistance in Madrid, Spain. J Clin Microbiol. 2010;48(10):3703-7. [View at Publisher] [DOI] [PMID] [Google Scholar]
11. Mansour KB, Burucoa C, Zribi M, Masmoudi A, Karoui S, Kallel L, et al. Primary resistance to clarithromycin, metronidazole and amoxicillin of Helicobacter pylori isolated from Tunisian patients with peptic ulcers and gastritis: a prospective multicentre study. Ann Clin Microbiol Antimicrob. 2010:9:22. [View at Publisher] [DOI] [PMID] [Google Scholar]
12. Klesiewicz K, Nowak P, Karczewska E, Skiba-Kurek I, Wojtas-Bonior I, Sito E, et al. PCR-RFLP detection of point mutations A2143G and A2142G in 23S rRNA gene conferring resistance to clarithromycin in Helicobacter pylori strains. Acta Biochim Pol. 2014;61(2):311-5. [View at Publisher] [DOI] [PMID] [Google Scholar]
13. Kargar M, Baghernejad M, Doosti A. Role of NADPH-insensitive nitroreductase gene to metronidazole resistance of Helicobacter pylori strains. Daru. 2010;18(2):137-40. [View at Publisher] [PMID] [Google Scholar]
14. Bauernfeind A. Comparison of the antibacterial activities of the quinolones Bay 12-8039, gatifloxacin (AM 1155), trovafloxacin, clinafloxacin, levofloxacin and ciprofloxacin. J Antimicrob Chemother. 1997;40(5):639-51. [View at Publisher] [DOI] [PMID] [Google Scholar]
15. Moore RA, Beckthold B, Wong S, Kureishi A, Bryan LE. Nucleotide sequence of the gyrA gene and characterization of ciprofloxacin-resistant mutants of Helicobacter pylori. Antimicrob Agents Chemother. 1995;39(1):107-11. [View at Publisher] [DOI] [PMID] [Google Scholar]
16. Gerrits MM, van Vliet AH, Kuipers EJ, Kusters JG. Helicobacter pylori and antimicrobial resistance: molecular mechanisms and clinical implications. Lancet Infect Dis. 2006;6(11):699-709. [View at Publisher] [DOI] [PMID] [Google Scholar]
17. Siavoshi F, Saniee P, Khalili-Samani S, Hosseini F, Malakutikhah F, Mamivand M, et al. Evaluation of methods for H. pylori detection in PPI consumption using culture, rapid urease test and smear examination. Ann Transl Med. 2015;3(1):11. [View at Publisher] [DOI] [PMID] [Google Scholar]
18. Momtaz H, Souod N, Dabiri H, Sarshar M. Study of Helicobacter pylori genotype status in saliva, dental plaques, stool and gastric biopsy samples. World J Gastroenterol. 2012;18(17):2105-11. [View at Publisher] [DOI] [PMID] [Google Scholar]
19. Glocker E, Stueger H-P, Kist M. Quinolone resistance in Helicobacter pylori isolates in Germany.Antimicrob Agents Chemother. 2007;51(1):346-9. [View at Publisher] [DOI] [PMID] [Google Scholar]
20. Mahmoudi L, Sharifzadeh F, Mousavi S, Pourabbas B, Niknam R. Susceptibility testing of Helicobacter pylori: Comparison of E-test and disk diffusion for metronidazole and mutations in rdxA gene sequences of Helicobacter pylori strains. Trends in Pharmaceutical Sciences. 2015;1(4):235-42. [View at Publisher] [Google Scholar]
21. Agudo S, Pérez-Pérez G, Alarcón T, López-Brea M. Rapid detection of clarithromycin resistant Helicobacter pylori strains in Spanish patients by polymerase chain reaction-restriction fragment length polymorphism. Rev Esp Quimioter. 2011;24(1):32-6. [View at Publisher] [PMID] [Google Scholar]
22. Vakil N, Megraud F. Eradication therapy for Helicobacter pylori. Gastroenterology. 2007;133(3):985-1001. [View at Publisher] [DOI] [PMID] [Google Scholar]
23. Ghotaslou R, Leylabadlo HE, Asl YM. Prevalence of antibiotic resistance in Helicobacter pylori: A recent literature review. World J Methodol. 2015;5(3):164-74. [View at Publisher] [DOI] [PMID] [Google Scholar]
24. Debets-Ossenkopp YJ, Pot RG, Van Westerloo DJ, Goodwin A, Vandenbroucke-Grauls CM, Berg DE, et al. Insertion of mini-IS 605 and deletion of adjacent sequences in the nitroreductase (rdxA) gene cause metronidazole resistance in Helicobacter pylori NCTC11637. Antimicrob Agents Chemother. 1999;43(11):2657-62. [View at Publisher] [DOI] [PMID] [Google Scholar]
25. Khademi F, Poursina F, Hosseini E, Akbari M, Safaei HG. Helicobacter pylori in Iran: A systematic review on the antibiotic resistance. Iran J Basic Med Sci. 2015;18(1):2-7. [View at Publisher] [PMID] [Google Scholar]
26. Abdollahi H, Savari M, Zahedi MJ, DARVISH MS, HAYATBAKHSH AM. A study of rdxA gene deletion in metronidazole resistant and sensitive Helicobacter pylori isolates in Kerman, Iran. Jundishapur J Microbiol. 2011;4(2):99-104. [View at Publisher] [Google Scholar]
27. Ramzy I, Elgarem H, Hamza I, Ghaith D, Elbaz T, Elhosary W, et al. Genetic mutations affecting the first line eradication therapy of Helicobacter pylori-infected Egyptian patients. Rev Inst Med Trop Sao Paulo. 2016;58:88. [View at Publisher] [DOI] [PMID] [Google Scholar]
28. Mirzaei N, Poursina F, Moghim S, Rahimi E, Safaei HG. The mutation of the rdxA gene in metronidazole-resistant Helicobacter pylori clinical isolates. Adv Biomed Res. 2014;3:90. [View at Publisher] [DOI] [PMID] [Google Scholar]
29. Thung I, Aramin H, Vavinskaya V, Gupta S, Park J, Crowe S, et al. the global emergence of Helicobacter pylori antibiotic resistance. Aliment Pharmacol Ther. 2016;43(4):514-33. [View at Publisher] [DOI] [PMID] [Google Scholar]
30. Zhao L-J, Huang Y-Q, Chen B-P, Mo X-Q, Huang Z-S, Huang X-F, et al. Helicobacter pylori isolates from ethnic minority patients in Guangxi: Resistance rates, mechanisms, and genotype. World J Gastroenterol. 2014;20(16):4761-70. [View at Publisher] [DOI] [PMID] [Google Scholar]
31. Khademi F, Sahebkar AH, Vaez H, Arzanlou M, Peeridogaheh H. Characterization of clarithromycin-resistant Helicobacter pylori strains in Iran: A systematic review and meta-analysis. J Glob Antimicrob Resist. 2017;10:171-8. [View at Publisher] [DOI] [PMID] [Google Scholar]
32. Karczewska E, Wojtas I, Budak A. Prevalence of Helicobacter pylori primary resistance to antimicrobial agents in Poland and around the world. Potepy Mikrobiologii. 2009;48(1):31-41. [View at Publisher] [Google Scholar]
33. Magraud F. Helicobacter pylori antibiotic resistance. Prevalence, importance and advance in testing. Gut. 2004;53(9):1374-84. [View at Publisher] [DOI] [PMID] [Google Scholar]
34. Francavilla R, Lionetti E, Castellaneta S, Margiotta M, Piscitelli D, Lorenzo L, et al. Clarithromycin-resistant genotypes and eradication of Helicobacter pylori. J Pediatr. 2010;157(2):228-32. [View at Publisher] [DOI] [PMID] [Google Scholar]
35. Sadeghifard N, Seidnazari T, Ghafourian S, Soleimani M, Maleki A, Qomi MA, et al. Survey in Iran of clarithromycin resistance in Helicobacter pylori isolates by PCR-RFLP. Southeast Asian J Trop Med Public Health. 2013;44(1):89-95. [View at Publisher] [PMID] [Google Scholar]
36. Abdollahi H, Savari M, Zahedi MJ, Moghadam SD, Abasi MH. Detection of A2142C, A2142G, and A2143G mutations in 23s rRNA gene conferring resistance to clarithromycin among Helicobacter pylori isolates in Kerman, Iran. Iran J Med Sci. 2011;36(2):104-10. [View at Publisher] [DOI] [PMID] [Google Scholar]
37. Eghbali Z, Mojtahedi A, Ansar MM, Asl SF, Aminian K. Detection of 23SrRNA mutations strongly related to clarithromycin resistance in Helicobacter pylori strains isolated from patients in the north of Iran. Jundishapur J Microbiol. 2016;9(2):e29694. [View at Publisher] [DOI] [PMID] [Google Scholar]
38. Suzuki RB, Lopes RAB, da Câmara Lopes GA, Ho TH, Sperança MA. Low Helicobacter pylori primary resistance to clarithromycin in gastric biopsy specimens from dyspeptic patients of a city in the interior of São Paulo, Brazil. BMC Gastroenterol. 2013:13:164. [View at Publisher] [DOI] [PMID] [Google Scholar]
39. Vazirzadeh J, Falahi J, Moghim S, Narimani T, Rafiei R, Karbasizadeh V. Molecular assessment of resistance to clarithromycin in Helicobacter pylori strains isolated from patients with dyspepsia by fluorescent in situ hybridization in the center of Iran. Biomed Res Int. 2020:2020:2304173. [View at Publisher] [DOI] [PMID] [Google Scholar]
40. Trespalacios-Rangél AA, Otero W, Arévalo-Galvis A, Poutou-Piñales RA, Rimbara E, Graham DY. Surveillance of levofloxacin resistance in Helicobacter pylori isolates in Bogotá-Colombia (2009-2014). PLoS One. 2016;11(7):e0160007. [View at Publisher] [DOI] [PMID] [Google Scholar]
41. Zaki M, Othman W, Ali MA, Shehta A. Fluoroquinolone-resistant Helicobacter pylori strains isolated from one Egyptian University Hospital: molecular aspects. J Microbiol Antimicrobial Agents. 2016;2(1):26-31. [View at Publisher] [Google Scholar]
42. Ontsira Ngoyi EN, Atipo Ibara BI, Moyen R, Ahoui Apendi PC, Ibara JR, Obengui O, et al. Molecular detection of Helicobacter pylori and its antimicrobial resistance in Brazzaville, Congo. Helicobacter. 2015;20(4):316-20. [View at Publisher] [DOI] [PMID] [Google Scholar]
43. Kipritci Z, Gurol Y, Celik G. Antibiotic Resistance Results of Helicobacter pylori in a University Hospital: Comparison of the Hybridization Test and Real-Time Polymerase Chain Reaction. Int J Microbiol. 2020:2020:8853298. [View at Publisher] [DOI] [PMID] [Google Scholar]
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |





 goums.ac.ir
goums.ac.ir yahoo.com
yahoo.com