Volume 19, Issue 1 (Jan-Feb 2025)
mljgoums 2025, 19(1): 27-31 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Panahi T, Asadpour L, Ranji N. Detection of β-lactamase genes and characterization of class 1 integrons in multidrug-resistant Pseudomonas aeruginosa strains in Guilan, Iran. mljgoums 2025; 19 (1) :27-31
URL: http://mlj.goums.ac.ir/article-1-1564-en.html
URL: http://mlj.goums.ac.ir/article-1-1564-en.html
1- Department of Biology, Rasht Branch, Islamic Azad University, Rasht, Iran
2- Department of Biology, Rasht Branch, Islamic Azad University, Rasht, Iran ,l.asadpour@yahoo.com
2- Department of Biology, Rasht Branch, Islamic Azad University, Rasht, Iran ,
Full-Text [PDF 514 kb]
(755 Downloads)
| Abstract (HTML) (2447 Views)
Results
Antibiotic resistance profile of P. aeruginosa strains
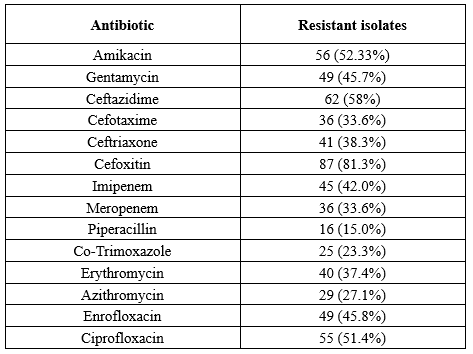
A total of 110 P. aeruginosa isolates were recovered during the study period. Based on antibiotic susceptibility testing, the highest phenotypic resistance was against cefoxitin and ceftazidime. Meanwhile, colistin and piperacillin were the most effective antibiotics. More than 85% of the isolates (n = 90) exhibited an MDR phenotype (Resistant to three classes of antibiotics), while 37% were classified as extensively drug-resistant (XDR) phenotype (Resistant to three classes of antibiotics plus imipenem). Seven isolates were resistant to all evaluated antibiotics. Moreover, according to the DDST, 40 isolates (37.4%) were ESBL-positive and 28 isolates (25.45%) were MBL-positive. The results of antibacterial resistance are presented in Table 2.
β-lactamase genes, including bla-VIM, IMP, SIM, GIM, SPM, OXA10, and OXA2, were detected in ESBL- and MBL-positive strains. Based on the results of PCR, the frequency of VIM, SIM, IMP, SPM, and OXA2 genes was 10.3%, 14%, 20.6%, 1.9%, and 4%, respectively. OXA 10, SPM, and GIM genes were not detected in any of the isolates. Furthermore, the int1 gene was identified in 37 isolates (34.6%). Int2 gene was not detected in any of the isolates.
Characterization of the int1 gene
Primers related to conserved regions of int1 were used for the detection of int1 polymorphism in P. aeruginosa strains. PCR results showed bands with lengths between 700 and 3000 bp (Figure 1).
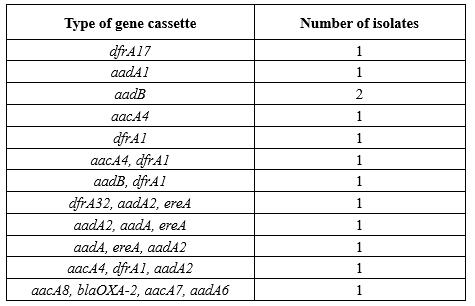
Among 13 selected isolates, 12 different types of gene cassettes in class 1 integrons and 18 different genes were identified in the sequence analysis of the int1 gene. The most frequent genes were dfrA1 (n = 5), aadA2 (n = 4), and aacA4 (n = 4). The class D β-lactamase gene, blaOXA-2, was the only bla gene identified in the examined isolates. The number of gene cassettes in tested isolates varied between 1 and 4. The type of gene cassettes identified in class 1 integrons among P. aeruginosa isolates is shown in Table 3.
Discussion
P. aeruginosa, one of the most common nosocomial infectious agents, has led to the death of a large number of hospitalized patients (15). ESBL antibiotics are commonly used for the treatment of diseases caused by these Gram-negative bacteria. However, some bacteria are resistant to β-lactam antibiotics due to ESBL production (16). In the current study, the rate of MDR P. aeruginosa was 85%. P. aeruginosa, especially its MDR strains, is very common in Iranian hospitals. Similar resistance rates were reported in other studies in Iran. For instance, Nikokar et al. documented a rate of 45.3% MDR among burn patients in Guilan in 2012 (17). In 2017, Fazeli et al. evaluated patients admitted to a teaching hospital and reported a rate of 73% MDR (18). In 2015, Ghanbarzadeh et al. conducted a single-center study on burn patients in Tehran and estimated the MDR rate at 93.1% (19). In our study, the highest and lowest rates of resistance belonged to cefoxitin (81.3%) and piperacillin (15%), respectively. Rates of resistance to ceftazidime and ceftriaxone were 62% and 56%, respectively. Moreover, our results suggested high resistance to imipenem at 42.05%, and the rate of resistance to cefotaxime was 33.6%. The increase in resistance to cephalosporins and imipenem is concerning. This might be due to selective pressure on these antibiotics, and it is highly important to monitor the prescription of these antibiotics. Following present results, Ameen et al. investigated the antibiotic resistance profile in P. aeruginosa strains and reported that among 230 strains of P. aeruginosa, 49.5% were resistant to imipenem (20).
In the present study, 37% of the isolates showed XDR phenotype, 37.4% of isolates were ESBL-positive, and 25.45% were MBL-positive based on DDST. The emergence of ESBL-producing P. aeruginosa is increasingly reported as a major cause of healthcare-associated infections (3). Overuse of antibiotics can lead to more resistant strains, resulting in gene and horizontal diffusion. High frequencies of ESBL production in P. aeruginosa isolates from burn patients in Iran were previously reported by Rafiee et al. (39.2% ESBL and 37.3% MBL production among 51 P. aeruginosa isolates) (21). This study demonstrated that the most prevalent genotype for MBL production was blaIMP, which was detected in 20.6% of isolates, followed by SIM (14%), VIM (10.3%), SPM (1.9%), and OXA2 (4%). OXA 10 and GIM were not observed in any of the strains. The blaVIM and blaIMP genes were common in MDR isolates of P. aeruginosa in previous studies in Iran (22,23).
On the other hand, a similar study in South Africa reported low frequency of blaIMP, while blaVIM was not detected among P. aeruginosa isolates (3). Moreover, in accordance with our results, blaIMP was the most frequently detected metallo-β-lactamase gene in a study conducted by Haghi et al. (9). The prevalence of class 1 integrons in this study was 34.6%, which is similar to the findings of Khosravi et al. in Ahvaz (24). The gene cassettes identified in 13 selected isolates included 1-4 different genes in a class 1 integron. A class 1 integron carrying the carbapenem resistance gene (aacA8-blaOxa-2-aadA7-aadA6) was also identified in meropenem-resistant P. aeruginosa. A high frequency of aminoglycoside resistance gene cassettes, including aadA family, which induce streptomycin and spectinomycin resistance, was identified in this assay. Gene cassettes dfrA1, aad8, dfrA17, dfrA32, and aacA8 were identified for the first time in this study.
The high frequency of aminoglycoside resistance gene cassettes in class 1 integrons has been reported in several studies. In a previous report, different aminoglycoside resistance genes, including aadA1, aadA2, aadA5, aadA6, aadB, accA4, and aac(6)-IIa, and three bla genes blaOXA10, blaP1, and blaCARB-8 were identified in integrons structure (25).
In a study conducted by Ahmadian et al. in northern Iran, the resistance genes identified in class I integrons included aadA6-orfD (35.71%), aacA4-blaOXA-10 (21.42%), aadB-aacA4-blaOXA-10 (19.04%), blaOXA-10-aacA4-VIM1 (11.9%), aacA4-catB10 (7.14%), aacA5-aadA1-cmlA5 (7.14%), blaOXA31-aadA2 (4.76%), and aac(3)-Ic-aacA5-cmlA5 (4.76%). The blaOXA-10-aacA4-VIM1 cassette array and aminoglycoside resistance genes were predominant (26). The highest frequency belonged to aadA2, which is similar to our findings. In another study from Iran, the analysis of integron cassette sequences indicated the presence of dfrA17, dfrA7, aadB, aadA1, and dhfr1-sat2 resistance gene cassettes among the isolates (9).
The predominant cassette arrangement described by Hsiao et al. among 11 different types of gene cassette arrays in P. aeruginosa isolates in southern Taiwan included aac(6')-II-catB2-aadA2, aac(6')-II-aadA2, aac(6')-II-catB2, aacA4-aadA15, aacC1-orfA-orfB-aadA1, cm1A-aadA1, catB3-blaOxA-10-aadA15, aacA4-catB8-aadA1, aadB-orfF1-aadA11, dfrB1, and dfrB4a-aacA4-aacA4-aadA1 (27). In 2018, Chowdhury et al. examined P. aeruginosa isolates with widespread antibiotic resistance in Australia and identified two genomic islands that carried transposons Tn6161 and Tn6163, respectively. A genomic island GI1 gene cassette blaGES-5-aacA4-gcuE15-aphA15 was also found in a class 1 integron, which confirms resistance to carbapenems and aminoglycosides (28). The presence of the aacA4 gene cassette in our study was similar to the findings of Chowdhury et al. (28). In addition, a high prevalence of aadA gene, which shows resistance to streptomycin/spectinomycin, was observed in a study conducted by Odumosu et al. Gene cassette identification showed the presence of aadA6-orfD and aadA13, respectively (29).
In the present study, the investigated class 1 integrons included 12 gene cassettes: aadA2, aadA6, era, aacA4, dfrA1, aad8, aacA8, blaoxa2, aacA7, dfrA17, dfrA32, and aadB. Gene cassettes dfrA1, aad8, dfrA17, dfrA32, and aacA8 were first identified in this study. In a study in China, many genes, including aadA1, aadA2, aadA5, aadA6, aadB, accA4, and aac(6′)-IIa in integron structures were associated with aminoglycoside resistance. Resistance genes that are often identified (aadA family) are adenyl transferase aminoglycoside genes that cause resistance to streptomycin and spectinomycin. In addition, three genes, i.e., blaOXA10, blaP1, and blaCARB-8, were identified within integrons (25). Gene cassettes aadB, accA4, aadA1, and aadA2 in the mentioned study are similar to those found in our study.
Conclusion
The high prevalence of class 1 integrons and resistance genes is a serious concern. It may jeopardize the control and treatment of Pseudomonas infections, increase the length of hospital stays, and raise mortality rates. Appropriate measures must therefore be taken to ensure the proper use of antibiotics in clinical and agricultural settings. In general, the results of the present study showed that class I integrons are widely distributed among P. aeruginosa strains isolated from hospitals in Guilan. They can be considered carriers of gene cassettes and can play an important role in the acquisition of MDR in this bacterium.
Acknowledgement
Support by the Islamic Azad University, Rasht Branch, is gratefully acknowledged.
Funding sources
None.
Ethical statement
Since we did not use any animal models or patients and only used isolates that were previously obtained from clinical samples in laboratories, we did not require ethical approval for this study; however, we confirm that the study complies with all regulations.
Conflicts of interest
All contributing authors declared no conflicts of interest.
Author contributions
Tahereh Panahi: Data curation, writing-original draft. Leila Asadpour: Writing, review and editing, supervision, and methodology. Najmeh Ranji: Methodology.
Data availability statement
The data utilized and analyzed in this study can be obtained from the corresponding author upon request.
Full-Text: (379 Views)
Introduction
Pseudomonas aeruginosa (P. aeruginosa) is a Gram-negative bacterium responsible for a wide variety of infections, including urinary tract infection, pneumonia, septicemia, wound infection, meningitis, and many life-threatening infections (1). Various mechanisms, such as β-lactamase production, target mutation, overexpression of output pumps, and reduced outer membrane permeability, are believed to be involved in the resistance of P. aeruginosa to antimicrobial agents (2). Due to the significance of β-lactam antibiotics in hospital use, various forms of β-lactamases, e.g., extended-spectrum β-lactamases (ESBLs), AmpC β-lactamases, and metallo-β-lactamases (MBL), have evolved. ESBLs are the major enzymes in bacteria that are resistant to various β-lactam antibiotics, including penicillins, cephalosporins, and carbapenems. However, they are inhibited by β-lactam inhibitors, such as clavulanic acid, sulbactam, and tazobactam (3,4). These enzymes break down the amide bond of the β-lactam ring and inactivate antimicrobial agents. ESBL genes, including blaSHV, blaTEM, and blaCTX-M, are encoded by plasmids and can transfer between bacterial species (5,6). These enzymes are highly prevalent in Enterobacteriaceae and P. aeruginosa strains. Mutations in genes encoding TEM and SHV enzymes lead to the production of ESBLs with an extended substrate catalytic activity, allowing for the breakdown of all cephalosporins, penicillins, and aztreonam (7). An increase in the frequency of ESBLs has been reported in recent years, but it varies in different geographical locations. Overall, clinical treatment failure occurs when inappropriate antimicrobial therapy is used to treat infections caused by ESBL-producing P. aeruginosa strains. Therefore, enabling laboratory technicians to correctly identify ESBL-producing P. aeruginosa isolates would lead to a better selection of antibiotics and improved infection outcomes (8,9). The growing resistance of P. aeruginosa to several antibiotics, as a result of excessive antibiotic administration, has resulted in high levels of antibiotic resistance and cross-resistance between antibiotics and the emergence of multidrug-resistant (MDR) forms of P. aeruginosa (10). Integrons are genetic elements that can carry multiple antibiotic resistance genes.
To date, four classes of integrons have been described in Gram-negative bacterial isolates. Three main classes of integrons have a 5′ conserved segment, including an intI gene encoding an integrase and an attI recombination site, but have distinct 3′ conserved segments (6). In class 1 integrons, the 3′ conserved segment includes three open reading frames (ORFs)-qacEΔ1, a deletion derivative of the antiseptic resistance gene qacE, the sul1 sulfonamide resistance gene, and ORF5 (Of unknown function), or int genes, as in Tn402. Gene cassettes are randomly integrated with the region between 3' and 5' of integrons. These cassettes are mobile gene elements that encode one or more antibiotic resistance genes and do not have a promoter sequence in their structure; thus, the expression of gene cassettes depends on the expression of integrons. Therefore, integrons act both as a gene expression vector and a natural cloning system (7). More than 80 different gene cassettes of class 1 integrons have been described and shown to confer resistance to a wide range of antiseptics, disinfectants, and antibiotics, such as β-lactams, fluoroquinolones, aminoglycosides, chloramphenicol, trimethoprim, streptothricin, rifampin, erythromycin, fosfomycin, and lincomycin (11).
Few studies have been conducted to investigate the prevalence of MDR strains of P. aeruginosa producing ESBL in Guilan province, northern Iran. A previous study in Guilan Province investigated the antimicrobial resistance, biofilm-forming ability, and virulence potential of P. aeruginosa isolated from burn patients, and the results showed that 72.2% (65/90) of P. aeruginosa isolates were MDR, and 55.5% (50/90) and 35.6% (32/90) were positive for ESBL and MBL production, respectively (12). The present study was conducted to investigate the frequency of antibiotic resistance, detection of β-lactamase, and characterization of gene cassettes carried by integrons in MDR P. aeruginosa strains in Guilan Province.
Methods
Bacterial isolates
This descriptive cross-sectional study was performed in 2021. A total of 110 P. aeruginosa isolates were collected from different hospitals in Rasht, northern Iran, and identified using standard microbiological methods. All clinical samples were inoculated on eosin methylene blue (EMB), MacConkey, and blood agar, and incubated at 37℃ for 24 hours. P. aeruginosa colonies were identified using microbiological methods.
Antibiotic susceptibility testing
Antibiotic susceptibility testing was performed by the Kirby-Bauer disc diffusion method according to the instructions of the Clinical and Laboratory Standards Institute (CLSI, 2020). Antimicrobial discs, including gentamicin (30 μg), amikacin (30 μg), ciprofloxacin (5 μg), co-trimoxazole (25 μg), piperacillin (100 μg), ceftazidime (30 μg), meropenem (10 μg), imipenem (10 μg), colistin (10 μg), azithromycin (30 μg), kanamycin (30 μg), erythromycin (30 μg), ceftriaxone (30 μg), and cefotaxime (30 μg) were used for initial screening of P. aeruginosa strains. The ATCC 27853 standard strain of P. aeruginosa was used as a control. ESBL production was determined by the double disk diffusion method using disks of ceftazidime (30 μg) and cefotaxime (30 μg) alone and in combination with clavulanic acid (10 μg) on Muller Hinton agar. A positive test result was defined as a ≥5 mm increase in the zone diameter compared to a disk without clavulanic acid. Moreover, an imipenem-EDTA double-disk synergy test was used for MBL enzyme production assay. Enhancements in the diameter of the zone of inhibition in IMP+EDTA in comparison with the IMP-only disks were considered to be MBL producers.
Detection of β-lactamase and integron genes
The DNA of P. aeruginosa isolates positive in the DDST was extracted using a DNA extraction kit as instructed by the manufacturer (Sigma-Aldrich, St Louis, MO). Subsequently, the frequency of β-lactamase genes (bla-VIM, IMP, AMPC, SIM, GIM, SPM, OXA10, and OXA2) and int1 and Int2 integron genes was determined in strains using PCR with specific primers as shown in Table 1. PCR was carried out in 25 µl PCR volumes containing 10 ng template DNA, 0.5 mM of dNTPs, 10 pm of each primer, and 1 µl of Taq DNA polymerase in 1x PCR buffer. DNA amplification was performed in a Mastercycler Personal thermal cycler (Eppendorf, Germany) based on cycling parameters described in Table 1. PCR products were analyzed in 1% agarose gel containing 25 μg of ethidium bromide in tris-ethylenediaminetetraacetic acid (EDTA) buffer, and the gel was photographed under an ultraviolet illuminator using a gel documentation system and confirmed by sequencing.
Detection of gene cassettes in int1
Gene cassettes within class 1 integrons were amplified using 5'-CS and 3'-CS primer pairs (Table 1). The PCR products were then sequenced and identified using the basic local alignment search tool (BLAST) for the gene cassette screening strategy. For PCR products longer than 700 bp, internal primers were designed based on the obtained sequences, and the internal parts of the integrons were amplified and subsequently sequenced.
Pseudomonas aeruginosa (P. aeruginosa) is a Gram-negative bacterium responsible for a wide variety of infections, including urinary tract infection, pneumonia, septicemia, wound infection, meningitis, and many life-threatening infections (1). Various mechanisms, such as β-lactamase production, target mutation, overexpression of output pumps, and reduced outer membrane permeability, are believed to be involved in the resistance of P. aeruginosa to antimicrobial agents (2). Due to the significance of β-lactam antibiotics in hospital use, various forms of β-lactamases, e.g., extended-spectrum β-lactamases (ESBLs), AmpC β-lactamases, and metallo-β-lactamases (MBL), have evolved. ESBLs are the major enzymes in bacteria that are resistant to various β-lactam antibiotics, including penicillins, cephalosporins, and carbapenems. However, they are inhibited by β-lactam inhibitors, such as clavulanic acid, sulbactam, and tazobactam (3,4). These enzymes break down the amide bond of the β-lactam ring and inactivate antimicrobial agents. ESBL genes, including blaSHV, blaTEM, and blaCTX-M, are encoded by plasmids and can transfer between bacterial species (5,6). These enzymes are highly prevalent in Enterobacteriaceae and P. aeruginosa strains. Mutations in genes encoding TEM and SHV enzymes lead to the production of ESBLs with an extended substrate catalytic activity, allowing for the breakdown of all cephalosporins, penicillins, and aztreonam (7). An increase in the frequency of ESBLs has been reported in recent years, but it varies in different geographical locations. Overall, clinical treatment failure occurs when inappropriate antimicrobial therapy is used to treat infections caused by ESBL-producing P. aeruginosa strains. Therefore, enabling laboratory technicians to correctly identify ESBL-producing P. aeruginosa isolates would lead to a better selection of antibiotics and improved infection outcomes (8,9). The growing resistance of P. aeruginosa to several antibiotics, as a result of excessive antibiotic administration, has resulted in high levels of antibiotic resistance and cross-resistance between antibiotics and the emergence of multidrug-resistant (MDR) forms of P. aeruginosa (10). Integrons are genetic elements that can carry multiple antibiotic resistance genes.
To date, four classes of integrons have been described in Gram-negative bacterial isolates. Three main classes of integrons have a 5′ conserved segment, including an intI gene encoding an integrase and an attI recombination site, but have distinct 3′ conserved segments (6). In class 1 integrons, the 3′ conserved segment includes three open reading frames (ORFs)-qacEΔ1, a deletion derivative of the antiseptic resistance gene qacE, the sul1 sulfonamide resistance gene, and ORF5 (Of unknown function), or int genes, as in Tn402. Gene cassettes are randomly integrated with the region between 3' and 5' of integrons. These cassettes are mobile gene elements that encode one or more antibiotic resistance genes and do not have a promoter sequence in their structure; thus, the expression of gene cassettes depends on the expression of integrons. Therefore, integrons act both as a gene expression vector and a natural cloning system (7). More than 80 different gene cassettes of class 1 integrons have been described and shown to confer resistance to a wide range of antiseptics, disinfectants, and antibiotics, such as β-lactams, fluoroquinolones, aminoglycosides, chloramphenicol, trimethoprim, streptothricin, rifampin, erythromycin, fosfomycin, and lincomycin (11).
Few studies have been conducted to investigate the prevalence of MDR strains of P. aeruginosa producing ESBL in Guilan province, northern Iran. A previous study in Guilan Province investigated the antimicrobial resistance, biofilm-forming ability, and virulence potential of P. aeruginosa isolated from burn patients, and the results showed that 72.2% (65/90) of P. aeruginosa isolates were MDR, and 55.5% (50/90) and 35.6% (32/90) were positive for ESBL and MBL production, respectively (12). The present study was conducted to investigate the frequency of antibiotic resistance, detection of β-lactamase, and characterization of gene cassettes carried by integrons in MDR P. aeruginosa strains in Guilan Province.
Methods
Bacterial isolates
This descriptive cross-sectional study was performed in 2021. A total of 110 P. aeruginosa isolates were collected from different hospitals in Rasht, northern Iran, and identified using standard microbiological methods. All clinical samples were inoculated on eosin methylene blue (EMB), MacConkey, and blood agar, and incubated at 37℃ for 24 hours. P. aeruginosa colonies were identified using microbiological methods.
Antibiotic susceptibility testing
Antibiotic susceptibility testing was performed by the Kirby-Bauer disc diffusion method according to the instructions of the Clinical and Laboratory Standards Institute (CLSI, 2020). Antimicrobial discs, including gentamicin (30 μg), amikacin (30 μg), ciprofloxacin (5 μg), co-trimoxazole (25 μg), piperacillin (100 μg), ceftazidime (30 μg), meropenem (10 μg), imipenem (10 μg), colistin (10 μg), azithromycin (30 μg), kanamycin (30 μg), erythromycin (30 μg), ceftriaxone (30 μg), and cefotaxime (30 μg) were used for initial screening of P. aeruginosa strains. The ATCC 27853 standard strain of P. aeruginosa was used as a control. ESBL production was determined by the double disk diffusion method using disks of ceftazidime (30 μg) and cefotaxime (30 μg) alone and in combination with clavulanic acid (10 μg) on Muller Hinton agar. A positive test result was defined as a ≥5 mm increase in the zone diameter compared to a disk without clavulanic acid. Moreover, an imipenem-EDTA double-disk synergy test was used for MBL enzyme production assay. Enhancements in the diameter of the zone of inhibition in IMP+EDTA in comparison with the IMP-only disks were considered to be MBL producers.
Detection of β-lactamase and integron genes
The DNA of P. aeruginosa isolates positive in the DDST was extracted using a DNA extraction kit as instructed by the manufacturer (Sigma-Aldrich, St Louis, MO). Subsequently, the frequency of β-lactamase genes (bla-VIM, IMP, AMPC, SIM, GIM, SPM, OXA10, and OXA2) and int1 and Int2 integron genes was determined in strains using PCR with specific primers as shown in Table 1. PCR was carried out in 25 µl PCR volumes containing 10 ng template DNA, 0.5 mM of dNTPs, 10 pm of each primer, and 1 µl of Taq DNA polymerase in 1x PCR buffer. DNA amplification was performed in a Mastercycler Personal thermal cycler (Eppendorf, Germany) based on cycling parameters described in Table 1. PCR products were analyzed in 1% agarose gel containing 25 μg of ethidium bromide in tris-ethylenediaminetetraacetic acid (EDTA) buffer, and the gel was photographed under an ultraviolet illuminator using a gel documentation system and confirmed by sequencing.
Detection of gene cassettes in int1
Gene cassettes within class 1 integrons were amplified using 5'-CS and 3'-CS primer pairs (Table 1). The PCR products were then sequenced and identified using the basic local alignment search tool (BLAST) for the gene cassette screening strategy. For PCR products longer than 700 bp, internal primers were designed based on the obtained sequences, and the internal parts of the integrons were amplified and subsequently sequenced.
|
Table 1. Nucleotide sequences of primers used in this study
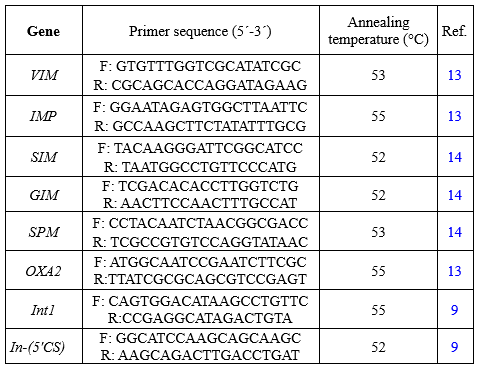 |
Results
Antibiotic resistance profile of P. aeruginosa strains
A total of 110 P. aeruginosa isolates were recovered during the study period. Based on antibiotic susceptibility testing, the highest phenotypic resistance was against cefoxitin and ceftazidime. Meanwhile, colistin and piperacillin were the most effective antibiotics. More than 85% of the isolates (n = 90) exhibited an MDR phenotype (Resistant to three classes of antibiotics), while 37% were classified as extensively drug-resistant (XDR) phenotype (Resistant to three classes of antibiotics plus imipenem). Seven isolates were resistant to all evaluated antibiotics. Moreover, according to the DDST, 40 isolates (37.4%) were ESBL-positive and 28 isolates (25.45%) were MBL-positive. The results of antibacterial resistance are presented in Table 2.
|
Table 2. Antimicrobial resistance properties in pseudomonas aeruginosa strains
 |
β-lactamase genes, including bla-VIM, IMP, SIM, GIM, SPM, OXA10, and OXA2, were detected in ESBL- and MBL-positive strains. Based on the results of PCR, the frequency of VIM, SIM, IMP, SPM, and OXA2 genes was 10.3%, 14%, 20.6%, 1.9%, and 4%, respectively. OXA 10, SPM, and GIM genes were not detected in any of the isolates. Furthermore, the int1 gene was identified in 37 isolates (34.6%). Int2 gene was not detected in any of the isolates.
Characterization of the int1 gene
Primers related to conserved regions of int1 were used for the detection of int1 polymorphism in P. aeruginosa strains. PCR results showed bands with lengths between 700 and 3000 bp (Figure 1).
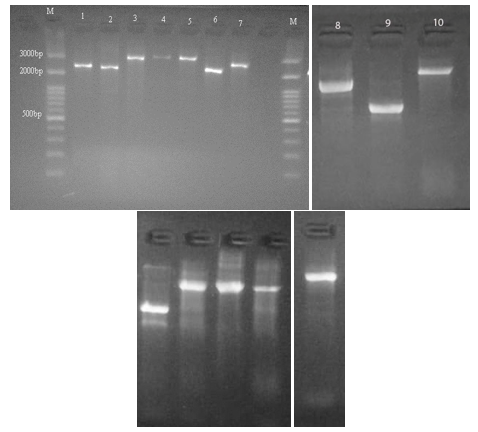 Figure 1. Size polymorphism in class 1 integron gene amplicons. Lanes M; 100bp molecular marker, Lanes 1-10; Size polymorphism in class 1 integron gene amplicons in tested isolates. |
Among 13 selected isolates, 12 different types of gene cassettes in class 1 integrons and 18 different genes were identified in the sequence analysis of the int1 gene. The most frequent genes were dfrA1 (n = 5), aadA2 (n = 4), and aacA4 (n = 4). The class D β-lactamase gene, blaOXA-2, was the only bla gene identified in the examined isolates. The number of gene cassettes in tested isolates varied between 1 and 4. The type of gene cassettes identified in class 1 integrons among P. aeruginosa isolates is shown in Table 3.
|
Table 3. Class 1 integron associated gene cassettes identified in pseudomonas aeruginosa strains
 |
Discussion
P. aeruginosa, one of the most common nosocomial infectious agents, has led to the death of a large number of hospitalized patients (15). ESBL antibiotics are commonly used for the treatment of diseases caused by these Gram-negative bacteria. However, some bacteria are resistant to β-lactam antibiotics due to ESBL production (16). In the current study, the rate of MDR P. aeruginosa was 85%. P. aeruginosa, especially its MDR strains, is very common in Iranian hospitals. Similar resistance rates were reported in other studies in Iran. For instance, Nikokar et al. documented a rate of 45.3% MDR among burn patients in Guilan in 2012 (17). In 2017, Fazeli et al. evaluated patients admitted to a teaching hospital and reported a rate of 73% MDR (18). In 2015, Ghanbarzadeh et al. conducted a single-center study on burn patients in Tehran and estimated the MDR rate at 93.1% (19). In our study, the highest and lowest rates of resistance belonged to cefoxitin (81.3%) and piperacillin (15%), respectively. Rates of resistance to ceftazidime and ceftriaxone were 62% and 56%, respectively. Moreover, our results suggested high resistance to imipenem at 42.05%, and the rate of resistance to cefotaxime was 33.6%. The increase in resistance to cephalosporins and imipenem is concerning. This might be due to selective pressure on these antibiotics, and it is highly important to monitor the prescription of these antibiotics. Following present results, Ameen et al. investigated the antibiotic resistance profile in P. aeruginosa strains and reported that among 230 strains of P. aeruginosa, 49.5% were resistant to imipenem (20).
In the present study, 37% of the isolates showed XDR phenotype, 37.4% of isolates were ESBL-positive, and 25.45% were MBL-positive based on DDST. The emergence of ESBL-producing P. aeruginosa is increasingly reported as a major cause of healthcare-associated infections (3). Overuse of antibiotics can lead to more resistant strains, resulting in gene and horizontal diffusion. High frequencies of ESBL production in P. aeruginosa isolates from burn patients in Iran were previously reported by Rafiee et al. (39.2% ESBL and 37.3% MBL production among 51 P. aeruginosa isolates) (21). This study demonstrated that the most prevalent genotype for MBL production was blaIMP, which was detected in 20.6% of isolates, followed by SIM (14%), VIM (10.3%), SPM (1.9%), and OXA2 (4%). OXA 10 and GIM were not observed in any of the strains. The blaVIM and blaIMP genes were common in MDR isolates of P. aeruginosa in previous studies in Iran (22,23).
On the other hand, a similar study in South Africa reported low frequency of blaIMP, while blaVIM was not detected among P. aeruginosa isolates (3). Moreover, in accordance with our results, blaIMP was the most frequently detected metallo-β-lactamase gene in a study conducted by Haghi et al. (9). The prevalence of class 1 integrons in this study was 34.6%, which is similar to the findings of Khosravi et al. in Ahvaz (24). The gene cassettes identified in 13 selected isolates included 1-4 different genes in a class 1 integron. A class 1 integron carrying the carbapenem resistance gene (aacA8-blaOxa-2-aadA7-aadA6) was also identified in meropenem-resistant P. aeruginosa. A high frequency of aminoglycoside resistance gene cassettes, including aadA family, which induce streptomycin and spectinomycin resistance, was identified in this assay. Gene cassettes dfrA1, aad8, dfrA17, dfrA32, and aacA8 were identified for the first time in this study.
The high frequency of aminoglycoside resistance gene cassettes in class 1 integrons has been reported in several studies. In a previous report, different aminoglycoside resistance genes, including aadA1, aadA2, aadA5, aadA6, aadB, accA4, and aac(6)-IIa, and three bla genes blaOXA10, blaP1, and blaCARB-8 were identified in integrons structure (25).
In a study conducted by Ahmadian et al. in northern Iran, the resistance genes identified in class I integrons included aadA6-orfD (35.71%), aacA4-blaOXA-10 (21.42%), aadB-aacA4-blaOXA-10 (19.04%), blaOXA-10-aacA4-VIM1 (11.9%), aacA4-catB10 (7.14%), aacA5-aadA1-cmlA5 (7.14%), blaOXA31-aadA2 (4.76%), and aac(3)-Ic-aacA5-cmlA5 (4.76%). The blaOXA-10-aacA4-VIM1 cassette array and aminoglycoside resistance genes were predominant (26). The highest frequency belonged to aadA2, which is similar to our findings. In another study from Iran, the analysis of integron cassette sequences indicated the presence of dfrA17, dfrA7, aadB, aadA1, and dhfr1-sat2 resistance gene cassettes among the isolates (9).
The predominant cassette arrangement described by Hsiao et al. among 11 different types of gene cassette arrays in P. aeruginosa isolates in southern Taiwan included aac(6')-II-catB2-aadA2, aac(6')-II-aadA2, aac(6')-II-catB2, aacA4-aadA15, aacC1-orfA-orfB-aadA1, cm1A-aadA1, catB3-blaOxA-10-aadA15, aacA4-catB8-aadA1, aadB-orfF1-aadA11, dfrB1, and dfrB4a-aacA4-aacA4-aadA1 (27). In 2018, Chowdhury et al. examined P. aeruginosa isolates with widespread antibiotic resistance in Australia and identified two genomic islands that carried transposons Tn6161 and Tn6163, respectively. A genomic island GI1 gene cassette blaGES-5-aacA4-gcuE15-aphA15 was also found in a class 1 integron, which confirms resistance to carbapenems and aminoglycosides (28). The presence of the aacA4 gene cassette in our study was similar to the findings of Chowdhury et al. (28). In addition, a high prevalence of aadA gene, which shows resistance to streptomycin/spectinomycin, was observed in a study conducted by Odumosu et al. Gene cassette identification showed the presence of aadA6-orfD and aadA13, respectively (29).
In the present study, the investigated class 1 integrons included 12 gene cassettes: aadA2, aadA6, era, aacA4, dfrA1, aad8, aacA8, blaoxa2, aacA7, dfrA17, dfrA32, and aadB. Gene cassettes dfrA1, aad8, dfrA17, dfrA32, and aacA8 were first identified in this study. In a study in China, many genes, including aadA1, aadA2, aadA5, aadA6, aadB, accA4, and aac(6′)-IIa in integron structures were associated with aminoglycoside resistance. Resistance genes that are often identified (aadA family) are adenyl transferase aminoglycoside genes that cause resistance to streptomycin and spectinomycin. In addition, three genes, i.e., blaOXA10, blaP1, and blaCARB-8, were identified within integrons (25). Gene cassettes aadB, accA4, aadA1, and aadA2 in the mentioned study are similar to those found in our study.
Conclusion
The high prevalence of class 1 integrons and resistance genes is a serious concern. It may jeopardize the control and treatment of Pseudomonas infections, increase the length of hospital stays, and raise mortality rates. Appropriate measures must therefore be taken to ensure the proper use of antibiotics in clinical and agricultural settings. In general, the results of the present study showed that class I integrons are widely distributed among P. aeruginosa strains isolated from hospitals in Guilan. They can be considered carriers of gene cassettes and can play an important role in the acquisition of MDR in this bacterium.
Acknowledgement
Support by the Islamic Azad University, Rasht Branch, is gratefully acknowledged.
Funding sources
None.
Ethical statement
Since we did not use any animal models or patients and only used isolates that were previously obtained from clinical samples in laboratories, we did not require ethical approval for this study; however, we confirm that the study complies with all regulations.
Conflicts of interest
All contributing authors declared no conflicts of interest.
Author contributions
Tahereh Panahi: Data curation, writing-original draft. Leila Asadpour: Writing, review and editing, supervision, and methodology. Najmeh Ranji: Methodology.
Data availability statement
The data utilized and analyzed in this study can be obtained from the corresponding author upon request.
Research Article: Research Article |
Subject:
Microbiology
Received: 2023/08/15 | Accepted: 2023/12/14 | Published: 2025/02/19 | ePublished: 2025/02/19
Received: 2023/08/15 | Accepted: 2023/12/14 | Published: 2025/02/19 | ePublished: 2025/02/19
References
1. Qin S, Xiao W, Zhou C, Pu Q, Deng X, Lan L, Liang H, Song X, Wu M. Pseudomonas aeruginosa: pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct Target Ther. 2022; 7(1): 199. [View at Publisher] [DOI] [PMID] [Google Scholar]
2. Pang Z, Raudonis R, Glick BR, Lin TJ, Cheng Z. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnology advances. 2019; 37(1): 177-92. [View at Publisher] [DOI] [PMID] [Google Scholar]
3. Hosu MC, Vasaikar SD, Okuthe GE, Apalata T. Detection of extended spectrum beta-lactamase genes in Pseudomonas aeruginosa isolated from patients in rural Eastern Cape Province, South Africa. Scientific reports. 2021; 11(1): 1-8. [View at Publisher] [DOI] [PMID] [Google Scholar]
4. Başkan C, Sırıken B, Tüfekci EF, Kılınç Ç, Ertürk Ö, Erol İ. Presence of quorum sensing system, virulence genes, biofilm formation and relationship among them and class 1 integron in carbapenem-resistant clinical Pseudomonas aeruginosa isolates. Arch Microbiol. 2022; 204(8): 464. [View at Publisher] [DOI] [PMID] [Google Scholar]
5. Büchler AC, Wüthrich D, Wicki Jauslin M, Egli A, Widmer AF. Nosocomial transmission of a blaVIM-2 carbapenemase integron between isolates of two different Pseudomonas species. Infect Control Hosp Epidemiol. 2022; 43(2): 245-247 [View at Publisher] [DOI] [PMID] [Google Scholar]
6. El Sayed Zaki M, Mostafa Mahmoud N, Anies Rizk M. Molecular Study of Integrase Gene I and Integrase Gene II in Clinical Isolates of Pseudomonas aeruginosa. Infect Disord Drug Targets. 2022; 22(7): 56-61. [View at Publisher] [DOI] [PMID] [Google Scholar]
7. Abd-Elmonsef MME, Maxwell SY. Class 1, 2 and 3 integrons in clinical Pseudomonas aeruginosa isolated from Tanta University Hospitals, Egypt. J Chemother. 2022; 34(4): 241-246. [View at Publisher] [DOI] [PMID] [Google Scholar]
8. Uddin F, Sohail M, Shaikh QH, Hussain MT, Roulston K, McHugh TD. Verona integron-encoded metallo-Beta-lactamase (VIM) and Vietnam extended-spectrum Beta-lactamase (VEB) producing Pseudomonas balearica from a clinical specimen. J Pak Med Assoc. 2022; 72(4): 761-763. [View at Publisher] [DOI] [PMID] [Google Scholar]
9. Haghi F, Keramati N, Hemmati F, Zeighami H. Distribution of Integrons and Gene Cassettes among Metallo-β-Lactamase Producing Pseudomonas aeruginosa Clinical Isolates. IEM. 2017; 3(2) :36-40. [View at Publisher] [Google Scholar]
10. Brovedan MA, Marchiaro PM, Díaz MS, Faccone D, Corso A, Pasteran F, et al. Pseudomonas putida group species as reservoirs of mobilizable Tn402-like class 1 integrons carrying blaVIM-2 metallo-β-lactamase genes. Infect Genet Evol. 2021; 96: 105131. [View at Publisher] [DOI] [PMID] [Google Scholar]
11. Aryanezhad M, Shakibaie M R, Karmostaji A, Shakibaie S. Prevalence of Class 1, 2, and 3 Integrons and Biofilm Formation in Pseudomonas aeruginosa and Acinetobacter baumannii among ICU and non-ICU Patients. IEM. 2016; 2(4): 1-7. [View at Publisher] [DOI] [Google Scholar]
12. Asadpour L. Antimicrobial resistance, biofilm-forming ability and virulence potential of Pseudomonas aeruginosa isolated from burn patients in northern Iran. J Glob Antimicrob Resist. 2018; 1(13): 214-220. [View at Publisher] [DOI] [PMID] [Google Scholar]
13. Garza-Ramos U, Morfin-Otero R, Sader HS, Rodriguez-Noriega A, Sanchez B, Carrillo S, Esparza-Ahumada, Silva-Sanchez J. Metallo-β-Lactamase Gene blaIMP-15 in a Class 1 Integron, In95, from Pseudomonas aeruginosa Clinical Isolates from a Hospital in Mexico, Antimicrob Agents Chemother. 2008; 52(8): 2943-2946. [View at Publisher] [DOI] [PMID] [Google Scholar]
14. Azimi A, Peymani A and Kianoush pour P. Phenotypic and molecular detection of metallo-β-lactamase-producing Pseudomonas aeruginosa isolates frompatients with burns in Tehran, Iran, Rev Soc Bras Med Trop. 2018; 51(5): 610-615. [View at Publisher] [DOI] [PMID] [Google Scholar]
15. Mirsalehian A, Feizabadi MM, Nakhjavani F A. Detection of VEB-1, OXA-10 and PER-1 genotypes in extended-spectrum-lactamase-producing Pseudomonas aeruginosa strains isolated from burn patients, February 2010Burns: journal of the International Society for Burn Injuries. 2010; 36(1):70-74. [View at Publisher] [DOI] [PMID] [Google Scholar]
16. Emami S, Nikokar I, Ghasemi Y, et al. Antibi-otic Resistance Pattern and Distribution of pslA Gene Among Biofilm Producing Pseudomonas aeruginosa Isolated From Waste Water of a Burn Center. Jundishapur J Microbiol. 2015; 8: e23669. [View at Publisher] [DOI] [PMID] [Google Scholar]
17. Nikokar I, Tishayar A, Flakiyan Z, Alijani K, Rehana-banisaeed S, Hossinpour M, et al. Antibiotic resistance and frequency of class 1 integrons among Pseudomonas aeruginosa, isolated from burn patients in Guilan, Iran. Iran J Microbiol 2013; 5(1): 36-41. [View at Publisher] [Google Scholar]
18. Fazeli H, Fatahi Bafghi M, Faghri M, Akbari R. Molecular study of PER and VEB genes is multidrug resistant Pseudomonas aeroginosa isolated from clinical specimens in Isfahan/Iran and their antibiotic resistance patterns. J Kerman Univ Med Sci. 2012; 19(4): 345-53. [View at Publisher] [Google Scholar]
19. Ghanbarzadeh Corehtash Z, Ahmad Khorshidi FF, Akbari H, Aznaveh AM. Biofilm formation and virulence factors among Pseudomonas aeruginosa isolated from burn patients. Jundishapur J Microbiol. 2015; 8: e22345. [View at Publisher] [DOI] [PMID] [Google Scholar]
20. Ameen N, Memon Z, Shaheen S, Fatima G, Ahmed F. Imipenem Resistant Pseudomonas aeruginosa: The fall of the final quarterback. Pak J Med Sci. 2015; 31(3): 561-5. [View at Publisher] [DOI] [PMID] [Google Scholar]
21. Rafiee R, Eftekhar F, Tabatabaei SA, Minaee Tehrani D. Prevalence of extended-spectrum and metallo-β-lactamase production in AmpC β-lactamase producing Pseudomonas aeruginosa isolates from burns. Jundishapur J Microbiol. 2014; 7(9):e16436. [View at Publisher] [DOI] [PMID] [Google Scholar]
22. Anvarinejad M, Japoni A, Rafaatpour N, et al. Burn patients infected with Metallo-Beta-Lac-tamase-producing Pseudomonas aeruginosa: multidrug-resistant strains. Arch Trauma Res 2014; 3: e18182. [View at Publisher] [DOI] [PMID] [Google Scholar]
23. Fallah F, Shams Borhan R, Gholinejad Z, Zahirnia Z, Adabiyan S, Sattarzadeh Tabrizi M, et al . Detection of blaIMP and blaVIM Metallo-Beta-Lactamases Genes in Pseudomonas aeruginosa Strains Isolated from Wound of Burnt Patients in Tehran Shahid Motahari Hospital during. 2011, Iran. Qom Univ Med Sci J. 2013; 7(5) :21-27 [View at Publisher] [Google Scholar]
24. Khosravi AD, Motahar M, Abbasi Montazeri E. The frequency of class1 and 2 integrons in Pseudomonas aeruginosa strains isolated from burn patients in a burn center of Ahvaz, Iran. PLoS ONE. 2017; 12(8): e0183061. [View at Publisher] [DOI] [PMID] [Google Scholar]
25. Gu B, Tong M, Zhao W, Liu G, Ning M, Pan S, et al. Prevalence and characterization of class 1 integrons among Pseudomonas aeruginosa and Acinetobacter baumannii isolates from patients in Nanjing, China. J Clin Microbiol 2007; 45(1): 241- 3. [View at Publisher] [DOI] [PMID] [Google Scholar]
26. Ahmadian L, Haghshenas M, Mirzaei B, et al. Distribution and Molecular Characterization of Resistance Gene Cassettes Containing Class 1 Integrons in Multi-Drug Resistant (MDR) Clinical Isolates of Pseudomonas aeruginosa, Mazandaran,Iran, Infection and Drug Resistance. 2020; 13: 2773-2781. [View at Publisher] [DOI] [PMID] [Google Scholar]
27. Hsiao K, Lee M, Peng C. Detection and characterization of class 1integron-associated gene cassettes from Pseudomonas aeruginosa isolates in southern Taiwan. Biomarkers and Genomic Medicine. 2014; 6: 74-78. [View at Publisher] [DOI] [Google Scholar]
28. Chowdhury V, Gunjal S, Mehta M. Antibiotic resistance patterns of Pseudomonas aeruginosa in a tertiary care hospital in Central India. International Journal of Medical Science and Public Health. Int J Med Sci Public Health. 2013; 2(2): 386-389. [View at Publisher] [DOI] [Google Scholar]
29. Odumosu BT, Adeniyi BA, Chandra R. Analysis of integrons and associated gene cassettes in clinical isolates of multidrug resistant Pseudomonas aeruginosa from Southwest Nigeria. Ann Clin Microbiol Antimicrob. 2013; 12: 29. [View at Publisher] [DOI] [PMID] [Google Scholar]
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |








 goums.ac.ir
goums.ac.ir yahoo.com
yahoo.com